
Interaction of de novo cholesterol biosynthesis and Hippo signaling pathway in ductal carcinoma in situ (DCIS)—comparison with the corresponding normal breast epithelium
Highlight box
Key findings
• Most genes involved in cholesterol biosynthesis were more expressed in ductal carcinoma in situ (DCIS) than in the corresponding histologically normal (HN) epithelium. This differential expression was associated with a substantial change in their correlation profile.
What is known and what is new?
• The mevalonate cascade, the core of cholesterol biosynthesis, contributes to the regulation of the Hippo signaling pathway providing the isoprenoids required for GTPase activation, the nuclear accumulation of the YAP/TAZ coactivator, and the subsequent activation of the TEA domain transcription factor (TEAD)-dependent gene transcription.
• Present findings corroborated the hypothesis that a dysfunctional mevalonate pathway concurs with DCIS development.
What is the implication, and what should change now?
• Due to its cholesterol-lowering properties, the safe and low-cost treatment with statins represents a promising strategy to contrast this metabolic dysfunction and DCIS progression.
Introduction
Ductal carcinoma in situ (DCIS) is an intraductal neoplastic proliferation of epithelial cells without infiltration of the basement membrane (1,2). Since the introduction of mammographic screening in the early 1980s, DCIS incidence has increased dramatically, and at present, it accounts for around 18–25% of newly diagnosed breast tumors (3). Although considered a non-obligate precursor to invasive breast cancer (IBC), if left untreated, about 50% of DCIS progress to IBC even after several decades from diagnosis. Indeed, a 30-year follow-up study showed that 36% of untreated low-grade DCIS progressed to IBC (4,5).
While great efforts have been made to elucidate the molecular journey leading DCIS to IBC with the aim of a more accurate definition of the risk to progress (6-8), less attention has been paid to the molecular and metabolic changes associated with the transformation of the normal mammary epithelium in DCIS. Filling this gap should be very important to identify the early events that trigger and drive this transformation and prevent them.
In the past two decades, accumulating evidence has revealed that Yes-associated protein (YAP) and its homologous transcriptional coactivator with PDZ-binding motif (TAZ) play an essential role in the regulation of many physiological and pathological processes, including tissue homeostasis and regeneration, fibrosis, inflammation, and tumorigenesis (9,10). In particular, YAP/TAZ coactivator proved to be involved in several steps of breast cancer development and progression, including tumor initiation and proliferation, metastatic spread, and drug resistance (11,12).
According to the canonical pathway, the activity of YAP/TAZ is regulated by the Hippo kinases cassette that consists of the serine/threonine kinase 3 (also known as STE20-like kinases MST1), the adaptor protein Salvador family WW domain-containing protein 1 (SAV1), the large tumor suppressor kinase 1 (LATS1) and MOB kinase activator 1A (MOAB1A) (13). As schematically depicted in Figure 1, STK kinase in complex with SAV1 directly phosphorylates MOB1A, which in turn, phosphorylates LATS (14). Once activated, LATS phosphorylates YAP/TAZ, thus hampering its migration into the nucleus and promoting its accumulation in the cytoplasm, where YAP/TAZ is degraded (15). Conversely, when unphosphorylated, YAP/TAZ migrates and accumulates into the nuclear compartment, where after binding to the TEA domain transcription factor (TEAD), promotes the transcription of several genes involved in cell proliferation, death, and migration.
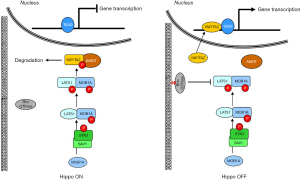
Although the Hippo kinases cascade has long been considered the major regulator of YAP/TAZ activity, novel regulatory mechanisms have recently been identified as able to inhibit YAP/TAZ phosphorylation, promote YAP/TAZ nuclear accumulation, and facilitate the interaction with TEAD. The best studied of these regulatory mechanisms is the small GTPase signaling pathway (16). In response to growth factors, acting as a master organizer of cytoskeleton dynamics and assembly of focal adhesions, the small GTPase pathway induces the accumulation of F-actin stress fibers, which, in turn, sequester angiomotin (AMOT), thereby promoting YAP/TAZ activation and nuclear translocation (17) (Figure 1). This mechanism was initially supposed as independent of the Hippo pathway, but further studies have demonstrated a link between small GTPase and Hippo pathways: the GTPase pathway can activate YAP/TAZ not only through the F-actin-mediated sequestration of the AMOT but also inhibiting the activity of LATS kinase (18).
To be effective, small GTPases require isoprenylation, a posttranslational modification that allows them to anchor cell membranes and be activated. The failure of small GTPases prenylation results in YAP/TAZ phosphorylation, its binding to AMOT, and degradation.
The main isoprenoids required for GTPases prenylation are farnesyl-pyrophosphate (FPP) and geranylgeranyl-pyrophosphate (GGPP), which are produced during the process leading to the de novo biosynthesis of cholesterol (Figure 2).
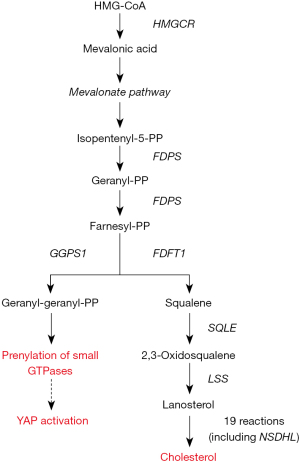
The accumulating clinical evidence that cholesterol metabolism is a risk factor for the onset and progression of breast cancer (19) and the experimental finding that cholesterol pathway genes were upregulated during breast cancer progression (20) stimulated the interest for a more accurate elucidation of the relationship among the production of isoprenoids during the cholesterol biosynthesis, Hippo kinase pathway, and YAP/TAZ activity in breast cancer initiation and progression.
In this context, in a very recent study (21), we have shown that in agreement with the hypothesis proposed by Sorrentino et al. (22), according to which the mevalonate cascade cooperates in the regulation of the YAP/TAZ activity promoting and sustaining mammary epithelial cells transformation and proliferation, we found that, in IBC, the progressive disruption of this regulatory mechanism associated with tumor grade and worse prognosis.
In the present study, we investigated whether such a disruption occurred already during the transformation of the normal mammary epithelium into DCIS. Therefore, we studied the relationship among the genes involved in the de novo cholesterol biosynthesis and those coding for the core components of the Hippo signaling pathway in a set of patient-matched samples of DCIS and corresponding histologically normal (HN) epithelium. We present this article in accordance with the MDAR reporting checklist (available at https://tbcr.amegroups.com/article/view/10.21037/tbcr-23-42/rc).
Methods
Dataset
For this study, we used a dataset publicly accessible at the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nhi.gov/geo/) through the accession number GSE14548, consisting of 18 breast cancer patient-matched samples of DCIS and corresponding HN epithelium. Despite the small number of samples, this dataset was the sole that allowed us to pursue our specific objective: to investigate the relationship between the genes involved in the de novo cholesterol biosynthesis and those coding for the core components of the Hippo signaling pathway during the transformation of the normal mammary epithelium into DCIS. Indeed, as described in the original article (23), each sample was processed by laser capture microdissection (LCM) to give highly enriched populations of normal or tumor epithelium. Conversely, in GSE16873, the only accessible dataset containing paired samples of DCIS and corresponding HN tissue, the specimens were epithelial and stroma compartments pooled together.
The original research (23) was approved by the Massachusetts General Hospital human research committee following National Institutes of Health human research study guidelines and deemed exempt from the patient’s informed consent as the samples are unidentifiable to the research team. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Gene set selection
Gene expression was measured using the Affymetrix whole-genome array U133X3P (GEO accession number GPL1352), and the gene expression estimates were provided as filtered and log2 transformed data. According to the aim of the study, we selected a panel of 16 genes that includes eight genes coding for the most important enzymes involved in cholesterol biosynthesis [3-hydroxy-3-methylglutaryl coenzyme-A (HMG-CoA) reductase (HMGCR), FDPS, FDFT1, GGPS1, SQLE, LSS, and NSDHL] and regulation (INSIG1); six coding the essential elements of the Hippo pathway [LATS1, MOB1A, SAV1, serine/threonine kinase 3 (STK3), WWTR1, and YAP1]; BIRC5 and CDK6, coding for two recognized direct downstream targets of the YAP-TEAD transcriptional complex, which play a crucial role, in the control of physiologic development of the mammary gland, respectively by coding the antiapoptotic survivin (24), and controlling the cyclin D-CDK4/CDK6 complex during cellular transition from the G1 into the S phase (25) (details on the selected genes in Table S1).
Statistical analysis
Since each gene can be recognized by different probes, thus resulting in numerous gene estimates, we calculated the mean value to obtain single expression data. Wilcoxon paired test was used to assess the differential expression of a given gene in DCIS and the corresponding HN tissue, and Spearman’s correlation coefficient was used to describe the correlation between genes. All analyses were performed using the open-source software R Core Team, version 4.1.2 (http://www.R-project.org), and a P value <0.05 was considered statistically significant.
Results
Expression of the genes involved in cholesterol biosynthesis in DCIS and corresponding HN tissue
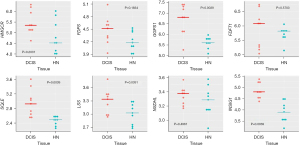
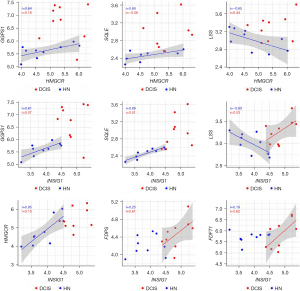
Wilcoxon paired test showed that all the genes involved in cholesterol biosynthesis were more expressed in DCIS when compared to the corresponding HN tissue, though only GGPS1, SQLE, LSS, and INSIG1 reached the statistical significance (Figure 3). The correlation analysis indicated that the differential expression of the genes was associated with a substantial change in their interrelationship. As shown in Figure 4, the genes more affected by this change were HMGCR, which codes for the rate-limiting enzyme in the mevalonate pathway, and INSIG1, which codes for the regulatory element of the SREBP-SCAP-Insig complex. Positively associated in HN tissue, HMGCR, and INSIG1 were almost unrelated in DCIS. Besides, their positive association with GGPS1 or SQLE considerably decreased in DCIS compared to the corresponding HN tissue, whereas their negative association with LSS switched to a positive one. In addition, INSIG1 was positively associated with FDPS or FDFT1 in DCIS but not in the corresponding HN tissue. Table S2 shows all the correlations among the genes involved in cholesterol biosynthesis.
Expression of the genes involved in the Hippo signaling pathway in DCIS and corresponding HN tissue and their association with those involved in cholesterol biosynthesis
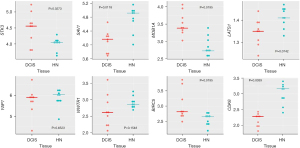
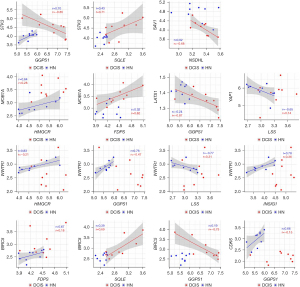
Wilcoxon paired test showed that, when compared with the corresponding HN tissue, all the genes coding for the essential elements of the Hippo signaling pathway were differentially expressed statistically in DCIS. The expression level of STK3 and MOB1A increased, whereas that of SAV1 and LATS1 decreased (Figure 5). As regards YAP1, WWTR1, BIRC5, and CDK6, while the expression level of YAP1 and WWTR1 did not significantly differ between DCIS and HN tissue, that of BIRC5 and CDK6 changed in a statistically significant manner: the expression level of BIRC5 increased while that of CDK6 decreased (Figure 5). The correlation analysis (Table S3) indicated that the positive association of STK3 with GGPS1 (r=0.70, P=0.0347) found in HN tissue switched to a negative one (r=−0.85, P=0.0061) in DCIS. Besides, it indicated: a considerable increase in the positive association of STK3 with SQLE as well as of the negative association of LATS1 with GGPS1, a decrease of the positive association of MOB1A with HMGCR, the appearance of a negative association between SAV1 and NSDHL (r=−0.68, P=0.0503), and a positive association between MOB1A and FDPS (r=0.62, P=0.0857) (Figure 6). As regards YAP1 and WWTR1, the negative association of YAP1 or WWTR1 with LSS found in NH tissue disappeared in DCIS as well as the positive association of WWTR1 with HMGCR, GGPS1, or INSIG1 (Figure 6).
Discussion
Present results indicated that most genes involved in the biosynthetic process leading to the de novo production of cholesterol were more expressed in the epithelial compartment of DCIS than the corresponding normal epithelium. The finding agrees with the clinical evidence that the HMG-CoA reductase (evaluated as immunohistochemical cytoplasmic staining) was moderately/strongly expressed in 70% of the assessed DCIS samples (26). Besides, the statistical analysis showed that they modified their interrelationship radically. In particular, HMGCR and INSIG1, which code, respectively, for the rate-limiting enzyme in the mevalonate cascade and the regulatory element of the SREBP-SCAP-Insig complex, lose the positive association they had in the HN tissue and substantially changed the relationship with the other genes of the biosynthetic pathway.
Given the role played by cholesterol in cell growth as an essential component in the assembly of new cell membranes and the pivotal function of HMG-CoA reductase and Insig in the regulation of cholesterol production, these results suggest a connection between the disruption of the mechanism that controls the biosynthetic process and the activation, in DCIS, of cell proliferation as indicated by the increased expression of BIRC5, which codes for the antiapoptotic survivin (24), and the substantial decrease in the expression level of CDK6, which codes for a cyclin-dependent kinase specifically involved in the negative control of the G1-phase of the cell cycle (27). To better understand the basis of such a connection, we remind that in normal tissue, the endogenous production of cholesterol is regulated by the sterol regulatory element-binding protein (SREBP), a membrane-bound transcription factor characterized by a sterol-sensing domain. In the presence of cholesterol, SREBP and its chaperone protein SCAP are retained in the endoplasmic reticulum membrane by the Insig protein. The binding of Insig to the SREBP/SCAP complex prevents the proteolytic generation of the transcriptionally active nuclear form of SREBP, thus limiting the transcription of the HMGCR gene and the activation of the downstream mevalonate cascade (28). In addition to the endoplasmic retention of the SREBP/SCAP complex, Insig recruits the HMG-CoA reductase present in the cytosol and promotes its degradation (29) as part of a tightly controlled feedback regulatory system that allows cells to synthesize cholesterol and essential nonsterol isoprenoids while avoiding their toxic overproduction (30).
The functional relationship between Insig and HMG-CoA reductase explains the very similar association profile of INSIG1 and HMGCR with the genes coding for the other component of the biosynthetic process that we found in HN tissue and, in particular, their inverse correlation with LSS, the gene coding the 2,3-oxidosqualene-lanosterol cyclase. Also known as lanosterol synthase, this enzyme catalyzes the formation of the sterol nucleus by the cyclization of (S)-2,3 oxidosqualene to lanosterol, which plays a crucial role in the Insig-mediated degradation of the HMG-CoA (31). The loss of the positive association between INSIG1 and HMGCR and the switch of their negative association with LSS to a positive one suggest that the dysfunctional activity of Insig possibly concurs with the development of a DCIS.
The possible causes leading to this dysfunction include a gene mutation in SCAP protein that makes the mutant SCAP resistant to the sterol-mediated conformational change and the binding to Insig much less efficient than the wild-type SCAP (28); epigenetic modifications that inhibit the correct translation of INSIG1 mRNA (32); posttranslational modifications, such as the Insig phosphorylation at Ser207, that reduce considerably the binding affinity of Insig to sterols or hamper the interaction with SCAP (33); the ubiquitin-dependent degradation of Insig promoted by the hypoxia-induced non-coding circular RNA (circINSIG1) (34). The most convincing of these causes is the microRNAs-mediated posttranscriptional modification of INSIG1 mRNA since cumulating evidence indicated that microRNA-122 inhibits the synthesis of specific INSIG1 isoform mRNAs by interfering with the usage of the promoter-proximal cleavage-polyadenylation site leading to the down-regulation of Insig protein (32).
Another interesting finding was the significant change in the correlation profile of GGPS1, the gene coding for geranyl-geranyl-PP synthase, which plays a crucial role in isoprenoids production. The positive association between GGPS1 and HMGCR or INSIG1 disappeared, whereas the positive association with SQLE, which codes for the enzyme that catalyzes the synthesis of squalene and the irreversible commitment to cholesterol, switched to a negative one in DCIS. When considered together, these results are of particular interest because they suggested that the disruption of the feedback mechanism that controls the mevalonate pathway lead to the abnormal synthesis of geranyl-geranyl-PP and consequently to increased production of the isoprenoids required for small GTPases anchoring and activation, which in turn, promote YAP nuclear translocation both by the F-actin-mediated sequestration of the AMOT and the inhibition of the LATS kinase activity (17,18). Noteworthy, the high negative association (r=−0.87) between the expression of GGPS1 and that of LATS1 suggests that, in addition to the GTPase-mediated inhibition of the LATS kinase activity, geranyl-geranyl-PP may exert negative feedback on the transcription of LATS1 gene. Once migrated into the nucleus, YAP can form a complex with TAZ and trigger the TEAD-mediated expression of the genes involved in cell proliferation and apoptosis, as supported by the decreased expression of CDK6 and the increased expression of BIRC5.
Of note is the increased expression of the STK3 and MOB1A genes, which code two crucial elements of the Hippo pathway, which appears as an effort to counteract, unsuccessfully, the transcriptional activity of the YAP/TAZ complex.
Unfortunately, the lack of other datasets with characteristics comparable to those of the dataset used in the present study—primarily the availability of epithelium-enriched paired samples of DCIS and corresponding healthy tissue—did not allow us to validate the results in an independent dataset. Indeed, when we interrogated the publicly accessible databases GEO and Array Express, we found several datasets containing DCIS samples and paired or independent IBC samples, very few datasets containing DCIS samples and independent samples of normal tissue, and only one dataset (GSE16873) containing patient-matched samples of DCIS and corresponding HN tissue. But, while the samples used in the present study were epithelial-enriched, those included in dataset GSE16873 contained both epithelial and stromal components that, contributing to a different extent to the gene expression level, would have led to misleading interpretations of the results. Conversely, although obtained in a small number of cases, present findings referred exclusively to the epithelial compartment providing insights on the disruption of the mechanism that regulates cholesterol biosynthesis occurring during the transformation of the normal mammary epithelium into DCIS and characterized by a decreased production of cholesterol, an increased production of isoprenoids and the subsequent promotion of YAP transcriptional activity.
Re-establishing the mechanism that controls cholesterol biosynthesis could represent a promising strategy to restore the correct equilibrium in the production of isoprenoids and cholesterol precursors and prevent the progression of DCIS to IBC due to the inhibition of the Hippo pathway activity. To achieve this, objective different strategies can be adopted, the most feasible of which is the inhibition of the activity of HMG-CoA reductase—the first rate-limiting specific enzyme of cholesterol synthesis—by statins and the activity inhibition of farnesyl pyrophosphate synthase—the regulatory enzyme crucial for the production of isoprenoids—by bisphosphonates. Currently used in clinical practice, respectively, to reduce breast cancer recurrence and mortality (35-37) and for treating patients with bone-related diseases, including osteoporosis and osteolytic bone metastases (38,39), statins have proven to inhibit the mevalonate pathway and isoprenoids production, thereby hampering the nuclear accumulation of YAP/TAZ complex and its transcriptional function (22) whereas, third-generation amino-bisphosphonates such as zoledronic acid and minodronate, have proved to interfere selectively with farnesyl pyrophosphate synthase thus preventing the biosynthesis of farnesyl pyrophosphate and the downstream geranylgeranyl pyrophosphate (40,41).
The combination of statins and bisphosphonates has been proposed to achieve a synergetic or multifunction effect in diseases related to the accumulation and/or persistence of prenylated proteins in cells (40). Even though no clinical studies have investigated the efficacy of such a combination in cancer disease, adding bisphosphonates to statins could help in counteracting the possible emergence of resistance to statin, as described in a meta-analysis of observational studies (42) and corroborated by the recent in vitro/in vivo study by Bhardwaj et al. (20) demonstrating that the dysregulation of a panel of genes involved in cholesterol biosynthesis, primarily HMGCR overexpression, significantly associated with resistance to statin treatment in mice.
Conclusions
Starting in the early 1980s, the increasing use of mammography screens has increased diagnosis of DCIS, especially among women >50 years of age, where it may represent up to 45% of all new cases of mammographically detected breast cancer (2,43). Although considered a non-obligate precursor to IBC, if left untreated, about 50% of DCIS progress to IBC even after several decades from diagnosis. Preventing such a progression is of paramount importance, and accomplishing this goal in a non-invasive manner represents a clinical challenge. Based on the cumulative evidence, corroborated by the present finding, that DCIS is associated with a dysfunctional mevalonate pathway leading to abnormal production of isoprenoids which in turn promote YAP activation, the safe and low-cost treatment with statins alone or in association with bisphosphonates could represent the winning strategy.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The author has completed the MDAR reporting checklist. Available at https://tbcr.amegroups.org/article/view/10.21037/tbcr-23-42/rc
Peer Review File: Available at https://tbcr.amegroups.org/article/view/10.21037/tbcr-23-42/prf
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at https://tbcr.amegroups.org/article/view/10.21037/tbcr-23-42/coif). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Leonard GD, Swain SM. Ductal carcinoma in situ, complexities and challenges. J Natl Cancer Inst 2004;96:906-20. [Crossref] [PubMed]
- Kuerer HM, Albarracin CT, Yang WT, et al. Ductal carcinoma in situ: state of the science and roadmap to advance the field. J Clin Oncol 2009;27:279-88. [Crossref] [PubMed]
- Bleyer A, Welch HG. Effect of three decades of screening mammography on breast-cancer incidence. N Engl J Med 2012;367:1998-2005. [Crossref] [PubMed]
- Collins LC, Tamimi RM, Baer HJ, et al. Outcome of patients with ductal carcinoma in situ untreated after diagnostic biopsy: results from the Nurses' Health Study. Cancer 2005;103:1778-84. [Crossref] [PubMed]
- Sanders ME, Schuyler PA, Simpson JF, et al. Continued observation of the natural history of low-grade ductal carcinoma in situ reaffirms proclivity for local recurrence even after more than 30 years of follow-up. Mod Pathol 2015;28:662-9. [Crossref] [PubMed]
- Schuetz CS, Bonin M, Clare SE, et al. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer Res 2006;66:5278-86. [Crossref] [PubMed]
- Tamimi RM, Baer HJ, Marotti J, et al. Comparison of molecular phenotypes of ductal carcinoma in situ and invasive breast cancer. Breast Cancer Res 2008;10:R67. [Crossref] [PubMed]
- Wiechmann L, Kuerer HM. The molecular journey from ductal carcinoma in situ to invasive breast cancer. Cancer 2008;112:2130-42. [Crossref] [PubMed]
- Zhao B, Ye X, Yu J, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 2008;22:1962-71. [Crossref] [PubMed]
- Zeng Q, Hong W. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell 2008;13:188-92. [Crossref] [PubMed]
- Zheng Y, Pan D. The Hippo Signaling Pathway in Development and Disease. Dev Cell 2019;50:264-82. [Crossref] [PubMed]
- Shi P, Feng J, Chen C. Hippo pathway in mammary gland development and breast cancer. Acta Biochim Biophys Sin (Shanghai) 2015;47:53-9. [Crossref] [PubMed]
- Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev 2013;27:355-71. [Crossref] [PubMed]
- Praskova M, Xia F, Avruch J. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr Biol 2008;18:311-21. [Crossref] [PubMed]
- Zhao B, Li L, Tumaneng K, et al. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev 2010;24:72-85. [Crossref] [PubMed]
- Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 1992;70:389-99. [Crossref] [PubMed]
- Mana-Capelli S, Paramasivam M, Dutta S, et al. Angiomotins link F-actin architecture to Hippo pathway signaling. Mol Biol Cell 2014;25:1676-85. [Crossref] [PubMed]
- Jang JW, Kim MK, Bae SC. Reciprocal regulation of YAP/TAZ by the Hippo pathway and the Small GTPase pathway. Small GTPases 2020;11:280-8. [Crossref] [PubMed]
- Nelson ER. The significance of cholesterol and its metabolite, 27-hydroxycholesterol in breast cancer. Mol Cell Endocrinol 2018;466:73-80. [Crossref] [PubMed]
- Bhardwaj A, Embury MD, Ju Z, et al. Gene signature associated with resistance to fluvastatin chemoprevention for breast cancer. BMC Cancer 2022;22:282. [Crossref] [PubMed]
- Coradini D, Ambrogi F, Infante G. Cholesterol de novo Biosynthesis in Paired Samples of Breast Cancer and Adjacent Histologically Normal Tissue: Association with Proliferation Index, Tumor Grade, and Recurrence-Free Survival: De novo cholesterol biosynthesis in breast tissue. Arch Breast Cancer 2023;10:187-99. [Crossref]
- Sorrentino G, Ruggeri N, Specchia V, et al. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat Cell Biol 2014;16:357-66. [Crossref] [PubMed]
- Ma XJ, Dahiya S, Richardson E, et al. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res 2009;11:R7. [Crossref] [PubMed]
- Martínez-Sifuentes MA, Bassol-Mayagoitia S, Nava-Hernández MP, et al. Survivin in Breast Cancer: A Review. Genet Test Mol Biomarkers 2022;26:411-21. [Crossref] [PubMed]
- Lucas JJ, Domenico J, Gelfand EW. Cyclin-dependent kinase 6 inhibits proliferation of human mammary epithelial cells. Mol Cancer Res 2004;2:105-14. [Crossref] [PubMed]
- Butt S, Butt T, Jirström K, et al. The target for statins, HMG-CoA reductase, is expressed in ductal carcinoma-in situ and may predict patient response to radiotherapy. Ann Surg Oncol 2014;21:2911-9. [Crossref] [PubMed]
- Nebenfuehr S, Kollmann K, Sexl V. The role of CDK6 in cancer. Int J Cancer 2020;147:2988-95. [Crossref] [PubMed]
- Yang T, Espenshade PJ, Wright ME, et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 2002;110:489-500. [Crossref] [PubMed]
- Jo Y, Lee PC, Sguigna PV, DeBose-Boyd RA. Sterol-induced degradation of HMG CoA reductase depends on interplay of two Insigs and two ubiquitin ligases, gp78 and Trc8. Proc Natl Acad Sci U S A 2011;108:20503-8. [Crossref] [PubMed]
- Luo J, Yang H, Song BL. Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol 2020;21:225-45. [Crossref] [PubMed]
- Song BL, Javitt NB, DeBose-Boyd RA. Insig-mediated degradation of HMG CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metab 2005;1:179-89. [Crossref] [PubMed]
- Norman KL, Chen TC, Zeiner G, et al. Precursor microRNA-122 inhibits synthesis of Insig1 isoform mRNA by modulating polyadenylation site usage. RNA 2017;23:1886-93. [Crossref] [PubMed]
- Xu D, Wang Z, Xia Y, et al. The gluconeogenic enzyme PCK1 phosphorylates INSIG1/2 for lipogenesis. Nature 2020;580:530-5. [Crossref] [PubMed]
- Xiong L, Liu HS, Zhou C, et al. A novel protein encoded by circINSIG1 reprograms cholesterol metabolism by promoting the ubiquitin-dependent degradation of INSIG1 in colorectal cancer. Mol Cancer 2023;22:72. [Crossref] [PubMed]
- Beckwitt CH, Brufsky A, Oltvai ZN, et al. Statin drugs to reduce breast cancer recurrence and mortality. Breast Cancer Res 2018;20:144. [Crossref] [PubMed]
- Bjarnadottir O, Romero Q, Bendahl PO, et al. Targeting HMG-CoA reductase with statins in a window-of-opportunity breast cancer trial. Breast Cancer Res Treat 2013;138:499-508. [Crossref] [PubMed]
- Iannelli F, Lombardi R, Milone MR, et al. Targeting Mevalonate Pathway in Cancer Treatment: Repurposing of Statins. Recent Pat Anticancer Drug Discov 2018;13:184-200. [Crossref] [PubMed]
- Räkel A, Boucher A, Ste-Marie LG. Role of zoledronic acid in the prevention and treatment of osteoporosis. Clin Interv Aging 2011;6:89-99. [PubMed]
- Lluch A, Cueva J, Ruiz-Borrego M, et al. Zoledronic acid in the treatment of metastatic breast cancer. Anticancer Drugs 2014;25:1-7. [Crossref] [PubMed]
- Sun S, McKenna CE. Farnesyl pyrophosphate synthase modulators: a patent review (2006 - 2010). Expert Opin Ther Pat 2011;21:1433-51. [Crossref] [PubMed]
- Drake MT, Clarke BL, Khosla S. Bisphosphonates: mechanism of action and role in clinical practice. Mayo Clin Proc 2008;83:1032-45. [Crossref] [PubMed]
- Undela K, Srikanth V, Bansal D. Statin use and risk of breast cancer: a meta-analysis of observational studies. Breast Cancer Res Treat 2012;135:261-9. [Crossref] [PubMed]
- Sakorafas GH, Farley DR, Peros G. Recent advances and current controversies in the management of DCIS of the breast. Cancer Treat Rev 2008;34:483-97. [Crossref] [PubMed]
Cite this article as: Coradini D. Interaction of de novo cholesterol biosynthesis and Hippo signaling pathway in ductal carcinoma in situ (DCIS)—comparison with the corresponding normal breast epithelium. Transl Breast Cancer Res 2023;4:26.


