
New-generation technologies for spatial tissue analysis, indispensable tools for deciphering intratumor heterogeneity in the development of antibody-drug conjugates and radio-immunoconjugates for cancer treatment
Introduction
Cancer treatment has been continuously improving over last 25 years, thanks to introduction into the clinic of targeted therapies which aim to specifically destroy malignant cells, with as less as possible damaging effects on normal tissues. These therapies either inhibit molecular anomalies crucial for survival of cancer cells or deliver the non-specific cell killing agents (such as DNA damaging agents, the basis of the standard chemo- or radiotherapy) specifically to the malignant cell deposits. Among the latter, antibody-drug conjugates (ADCs) or radio-immunoconjugates (RICs), consisting of an as much as possible cancer-specific antibody linked to an agent with cell killing properties (a chemotherapeutic or a radiation-emitting agent), have shown very encouraging results in last decade, to the point that some of them will change the treatment paradigm of several solid tumors (1). However, the response to ADCs varies among patients, highlighting the need for improvement in patient selection.
One of the much-evoked reasons for failure of any anticancer therapy, and in particular of targeted therapies, is intratumor heterogeneity (ITH). It covers a large variety of features, from the expression of the target molecule to the expression of numerous factors proper to the malignant cells or their benign neighborhood, which can impact the efficiency of targeted treatment. ITH has been considered even as the pillar of cancer resistance to treatment, with multiple facets that need to be assessed (2). It must be underlined, though, that has been intriguing pathologists and other tissue specialists for a long time. In the attempt to generate answers, for decades only histological, cytochemical, and immunohistochemical approaches have been used. Although modest in terms of the number of molecules simultaneously assessable in a tissue section (up to 5, in the best immunohistochemistry (IHC) protocols for automated multiplex staining), works published during the 20th century have undoubtfully demonstrated that malignant tumors display spatial ITH, provoked either by heterogeneity in tumor cellular composition or by heterogeneity in molecular anomalies within tumor tissue, or by both. Furthermore, ITH has been shown to increase practically after any cancer treatment, due to the treatment-induced changes both in the cellular and the acellular part of tumor tissue. Finally, after almost a century of focus to tumor cells as the main source of ITH, the 21st century shed the light on another very heterogeneous area within tumors, the tumor stroma, constituted of benign cells, extracellular matrix, and a plethora of secreted molecules which ensure intratumor communication as well as the “communication to the external worlds”, the future sites of metastasis.
In personalized oncology, the current century has also been a “Time of One to Many”: the concept “One Molecule—One Therapeutic Target—One Biomarker (of response)” has evolved into awareness that a powerful anticancer treatment must be based on several anticancer principles, and the predictive biomarkers must be syntheses of quantifications of many molecular alterations. This multi-element aspect is the main theme of the on-going evolution of biomarkers in medical and radiation oncology, allowed by great technological advances in molecular tissue analysis which enabled not only the “bulk tissue” high-plex molecular assessments but, most impressively, the integrative “omics” of tissue regions, cell groups or single cells. Thus, we have nowadays technologies capable of assessing a high number of molecular alterations and to associate them, without tissue lysis, to specific tumor tissue areas or cellular subpopulations. These approaches are known under name of spatial tissue analysis or spatial omics. Although still in early infancy, these methods represent one of the revolutions in oncology and biology in general, providing an unprecedented insight into how tissues are organized, and how this structural organization impacts tissue and organ function, in health and in disease.
We will review here the currently available technologies for tissue analysis with spatial resolution, their utility in biomarker research and development of anticancer treatments, in particular of ADCs, to finish by the perspectives in this exciting new field.
Spatial tissue analysis technologies based on image analysis
These technologies exploit two main approaches: (I) multiplex immunofluorescence (mIF) combined with analyses of digitalized images of stained tissue sections; or (II) immunostaining with metal-tagged antibodies, measurements performed by mass spectrometry (MS).
The fluorescent multiplexing is based on direct or indirect immunofluorescence (IF). To enable detection of low-abundant proteins, most IF (as well as chromogenic IHC) methods exploit the small molecule-based amplification technologies, among which tyramide-based signal amplification (TSA) is considered as one of the most powerful. TSA uses secondary antibodies conjugated to horseradish peroxidase which converts tyramide into an oxidized product that binds to tyrosine residues on or near the protein of interest. Multiple rounds of staining can be done, allowing detection of several proteins. The amplified signal is visualized by fluorescence detection which is significantly more sensitive in comparison to the detection of colored precipitate as in chromogenic IHC; for that reason, mIF is capable of detecting a much higher number of proteins in a tissue than the standard IHC.
Two most advanced currently available mIF-based technologies are PhenoCycler (formerly CODEX) and PhenoImager (formerly Phenoptics), which can be used together as PhenoCycler®-Fusion (Akoya Biosciences, Marlborough, MA, USA), and COMETTM (Lunaphore, Tolochenaz, CH, USA).
Spatial tissue analysis system developed by Akoya Biosciences combines a 9-color mIF tissue staining (OpalTM multiplex IHC kit: one antibody for nuclear staining and up to 8 antibodies for membranous proteins) optimized for reliable spectral unmixing (3,4). Tissue immunostaining can be performed manually or automatically, and results in a complex image that is further processed in an image analyzer that exploits a proprietary multispectral imaging technology (Phenoptics, which may contain the Mantra, Vectra 3, or Vectra Polaris Quantitative Pathology Imaging System). The analyzer’s software (inFormTM) can separate and quantify even weakly expressed and spectrally overlapping proteins within tissue compartments. In addition, automated trainable algorithms are used for tissue segmentation and derivation of IF-labeled cells’ phenotype. Plexing and capacity are strongly can go up to 100 protein biomarkers and 300+ slides per week, in a touchless automated fashion (adapted from PhenoCycler®-Fusion Product Brochure; the PhenoCycler®-Fusion Solution, application note: https://www.akoyabio.com/phenocycler-fusion-system/100-plex-app-note/). Figure 1 shows an example of tissue image obtained after tissue labeling by mIF using the Akoya system.
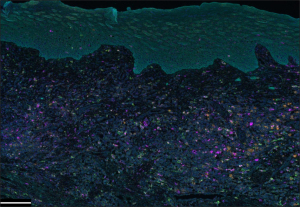
Lunaphore’s COMETTM is all-in-one staining and imaging platform composed of a microfluidics-based staining system and a tissue imager which allows visualization of up to 40 protein markers in a single sample. The platform has an integrated microscope which enables direct validation of the imaging setup and the automation of sequential data extraction. Data can be immediately evaluated through the Lunaphore Viewer software. If required, more in-depth image analysis can be carried out with the common image analysis tools that are compatible with the COMETTM platform (e.g., QuPath, HaloTM, Visiopharm, and ImageJ) (adapted from COMETTM Grant Application Support Package, https://lunaphore.com/resource-center/?category=Product%20Brochures).
Besides the mentioned two platforms, several other providers offer mIF-based mid-plex spatial tissue analysis, as shown in Table 1. In addition, using antibodies and visualization imaging from providers of their own choice, academic researchers develop mIF protocols that can evaluate up to 20 markers (6,11).
Table 1
| Principle | Type of detected target, number of targets detectable in one tissue section | Maximal surface of analysis | Spatial resolution/capacity for SC analysis | Apparatus/manufacturer |
|---|---|---|---|---|
| Technologies based on image analysis | ||||
| IHC, sequential | Proteins, ≤5 | Whole tissue section | 4–5 µm/weak | Benchmark Discovery/Ventana, Dako Omnis/Agilent, Bond Rx/Leica, UltraPLEX/Cell IDx |
| IF, sequential | Proteins, ≤65 | Whole tissue section except COMETTM: ROIs of 9 mm × 9 mm | 200 nm/weak | CellDIVE/Leica, CyCIF (5), 4i, UltraPLEX/Cell IDx, COMETTM/Lunaphore, ISP, Immuno-SABER, FlexVUETM/Ultivue, MACSimaTM/Miltenyi, BLEACH&STAIN (6) |
| IF, simultaneous | Proteins, ≤100 | Whole tissue section | 250 nm/moderate SC: OrionTM | PhenoCycler®-Fusion/Akoya, OrionTM/Rarecyte® |
| IMC | Proteins, ≤50 | ROIs of 300 µm × 300 µm max | 1 µm/moderate | Hyperion/Fluidigm and Standard Bio Tools |
| MIBI | Proteins, ≤100 | Whole tissue section | 400 nm/weak | MIBITM/IonPath |
| Technologies not based on image analysis | ||||
| IF simultaneous (orientation) + barcoded oligonucleotides cleaved by UV light | Proteins (~150), mRNA (18,000+, whole transcriptome) | Proteins: ROIs of 0.125 mm2 (50 cells minimum); mRNA: ROIs of 0.03 mm2 (200 cells minimum) | 200 nm/no | GeoMx® Digital Spatial Profiler/NanoString Technologies |
| IF simultaneous (orientation) + barcoded oligonucleotides cleaved by UV | Proteins (~125), mRNA (≥6,000) | ≤100 mm2 | <100 nm/strong | CosMxTM Spatial Molecular Imager/NanoString Technologies |
| MERFISH | mRNA panels of 140, 300, 500 | Whole tissue section | 100 nm/strong | MerscopeTM/Vizgen |
| Gene arrays | mRNA (18,000+, whole transcriptome) | ROIs of 11 mm × 11 mm | 55 µm/yes, with an external software | Xenium, Visium/10× Genomics |
Slide-seq, XYZseq, HDST, Stereo-seq, ZipSeq, Pick-seq, DBiT-seq, Seq-Scope, FISSEQ, STARmap, Seq-FISH+ [details: Hsieh et al. (7)]. Details of spatial omics technologies use in SC analysis can be referred to (8-10). SC, single-cell; IHC, immunohistochemistry; IF, immunofluorescence; ROI, region of interest; IMC, imaging mass cytometry; MIBI, multiplexed ion beam imaging; UV, ultraviolet; mRNA, messenger RNA; HDST, high-definition spatial transcriptomics; FISH, fluorescence in situ hybridization.
Two main available metal tag-based spatial technologies are imaging mass cytometry (IMC) and multiplexed ion beam imaging (MIBI).
IMC combines laser tissue ablation and mass cytometry by time-of-flight (CyTOF) (12). It can analyze up to 100 markers on a single tissue section, however, most of the currently developed panels contain no more than 50 markers. The immunostaining is performed by antibodies labeled with highly pure metal isotopes (instead of a fluorophore, in mIF), and followed by choosing the regions of interest (ROIs) for analysis by Hyperion Imaging System (Fluidigm, San Francisco, CA, USA). The tissue within ROIs is ablated by a laser pulse and vaporized to release metal particles (antibody ‘tags’) which are transferred to a CyTOF detector for quantification. This way the discrete signals from each metal tag are detected based on differences in mass instead of wavelength, at 1 Da resolution. The process of using metal tags with a defined atomic mass significantly reduces signal overlap, allowing much higher multiplexing than mIF imaging. Another important advantage is that the use of nonbiological metal isotopes essentially eliminates tissue background signals. The IMC exploiting Hyperion-based image analysis can also provide subcellular protein expression analysis (adapted from Hyperion Imaging System Product Brochure, Fluidigm, https://www.standardbio.com/products/instruments/hyperion#resources-anchor).
MIBI (IonPath Inc., Menlo Park, CA, USA) uses metal-chelator tags attached to antibodies as in the IMC technique; the difference lies in the sample analysis, which in the MIBI technique is based on time-of-flight secondary ion MS (TOF-SIMS) (13). A tissue section is ablated using an oxygen primary-ion beam, which releases metal isotopes from antibodies as secondary ions. The liberated particles are transported to a TOF mass spectrometer, which assigns them to distinct targets based on their atomic mass detection. That way each unique metal ion represents a protein. MIBI allows detection of 40–50 protein maximum, however with high-resolution images, and the antibody panels are rapidly increasing in number. This technology is still more used in the USA/Canada than in the European Union (EU) (adapted from https://www.ionpath.com/).
Spatial tissue analysis technologies not based on image analysis
These technologies utilize tissue section images only for orientation in ROI selection. However, they can generate images composed of digital signals associated with detected tissue molecules. Most of them provide a much higher plexing than the technologies based on stained slide image analysis. High-plex measurements of messenger RNA (mRNA) or protein abundance are enabled by oligonucleotide-based probe tags as well as by dedicated tag detection systems, proper to each technology. Several technologies can spatially evaluate the entire transcriptome (14-16) whereas the number of proteins detectable in one tissue section goes up to a few hundred. Spatial proteogenomic (17,18) as well as epigenomic-transcriptomic assays (19) are under intense development.
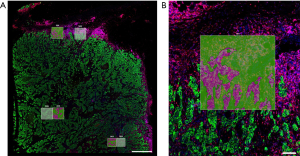
NanoString Technologies (Seattle, WA, USA) uses its proprietary probe barcoding system (nCounter® technology) and rigorously validated protein and mRNA probes, in two instruments dedicated to spatial tissue analysis, GeoMx® Digital Spatial Profiler (DSP) and CosMxTM Spatial Molecular Imager (SMI). Tissue labeling is performed by the IHC/in situ hybridization (ISH) principle (RNAScopeTM for ISH). After this step, which allows up to 5-plex mIF visualization of tissue sections on glass slides, the DSP scans the slides and presents a high-quality image for selection of ROIs. The oligonucleotide barcoded tags are released from user-selected ROIs with focused ultraviolet (UV) light. Released tags are quantitated with nCounter® or an Illumina® next-generation sequencer. Finally, the barcode reading output data are mapped back to the tissue location by an adapted software, resulting in a spatially resolved digital profile of mRNA or protein abundance. The instrument also contains a spatial data analysis (sDA) software which can offer ready-to-publish graphical presentations of the results. The DSP allows several types of ROI: geometric or free shapes, layers, cell-type ROIs, as well as ROI segmentation on two or more areas of analysis [e.g., tumor cells and tumor microenvironment (TME), Figures 2,3]. Thus, the DSP is an instrument of choice for exploration of tissue areas, however without capacity for real single-cell analysis. The latter is enabled by the SMI, which is an integrated system with cyclic ISH chemistry, an ultra-high-resolution imaging readout instrument enabling tissue segmentation as precise as one cell per ROI, and an interactive sDA and visualization software. Images created by the SMI are formed of spots, each corresponding to one mRNA or protein within a tissue. sDA capabilities include basic modules for quality control, normalization, cell typing, and spatial clustering, as well as advanced modules (in on-going development) for differential expression, cell proximity, signaling pathway analysis, and ligand-receptor co-expression. At the moment, the SMI allows simultaneous measurement of up to 6,000 mRNAs and more than 100 proteins at the single-cell level (adapted from GeoMx® DSP Grant Support Package, CosMxTM SMI Grant Support Package, https://nanostring.com/support/additional-resources/grant-support/).
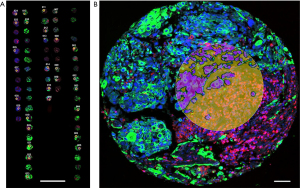
MERSCOPETM platform (Vizgen, Cambridge, MA, USA) utilizes the MERFISH technology which expands the capabilities of single molecule fluorescence ISH (FISH) by using combinatorial labeling, sequential imaging, and error-robust barcoding to detect RNA with sub-micron accuracy. Tissue sections are labeled by three antibodies for ROI selection, and mRNAs are detected by panels containing 140–500 gene probes. The visualizing software creates two-dimensional (2D) or three-dimensional (3D) tissue images from digital “dots” corresponding to each mRNA. MERSCOPETM doesn’t require sequencing, the measurements correlate with bulk RNA sequencing (RNA-seq) data, are highly reproducible between replicates and the detection efficacy is higher than in array-based platforms, identifying 70× more transcripts per gene (adapted from MERSCOPETM Product Brochure, https://vizgen.com/products/).
10× Genomics (San Francisco, CA, USA) provides two instruments for high-plex in situ molecular tissue analysis, Xenium and Visium. The technology behind is an array-based RNA capture, meaning that a tissue section is put on a special glass slide containing RNA probes instead of putting probes in solution on a glass slide with a tissue section. In Xenium, ligation of the probes generates a circular DNA probe which is enzymatically amplified and bound with fluorescent oligos, creating a bright, easy-to-image signal that has a high signal-to-noise ratio. The next step is generation of an optical signature, enabling identification of the target gene. Finally, a spatial map of the transcripts is built across the entire tissue section, using an integrated software. Visium is a higher-plex, bigger-surface (up to 11 mm × 11 mm) spatial tissue analyzer than Xenium, which can assess the whole transcriptome as well as perform protein abundance measurements. It utilizes barcoding of the RNA probes so each capture area has thousands of barcoded spots containing millions of capture oligonucleotides with spatial barcodes unique to that spot. IF is used to label proteins for tissue visualization. After post-capture tissue processing, the barcoded molecules are pooled to generate a library that is sequenced using common next-generation sequencing (NGS) equipment (adapted from Xenium Product Sheet, Visium Product Sheet, https://www.10xgenomics.com/).
Table 1 synthesizes characteristics of the described equipment.
Spatial tissue analysis and new anticancer drugs: ADCs and immunotherapy
High-plex molecular tissue analyzers offer an unprecedent help in answering many complex scientific questions in the fields of developmental biology, oncology (medical, radiation, immuno-oncology), cell therapy, neuroscience, dermatology, aging, and degenerative disease research, etc. In cancer treatment personalization, these instruments can reveal tumor tissue areas and/or cell subpopulations which are responsible for good or poor response to anticancer drugs. The number of publications of such results is growing, however a great part of them present “atlasing” of molecular expression in cancer tissue before and after an anticancer treatment; the discovery of biomarkers based on spatial tissue analysis is still slow, due to the relatively high cost of spatial technologies and the need to exploit them on larger cohorts of patient samples, collected in a controlled manner. However, several interesting observations have been reported, encouraging the use of these technologies in detailed exploration of tissue samples in translational cancer research. Rimm’s team was among the first to use NanoString’s DSP in discovery of predictive biomarker for immune checkpoint inhibitors (ICIs). They used a 44-protein panel and the cell-specific type of ROI segmentation to show that programmed death-ligand 1 (PD-L1) expression in CD68-positive cells (macrophages) and not in tumor cells is predictive and prognostic biomarker in melanoma treated by ICIs (20). A very recent paper from the MD Anderson Cancer Center reported on utilization of the same technology, with a 73 immune-related protein probe panel and spatial transcriptomics, in discovery of recurrence biomarkers of localized squamous cell anal cancer treated by chemoradiation. No differences in mRNA expression between recurring and non-recurring samples were observed, whereas recurring tumors had higher expression of FoxP3, BRAF, p38-MAPK, and phospho-Akt proteins in tumor cells, together with higher expression of PD-1, OX40L, and LAG3 in the TME (21).
Human epidermal growth factor receptor 2 (HER2)-expressing breast cancer is one of the best-known examples of high inter-tumor heterogeneity and ITH. ITH of HER2 protein expression as well as of genomic anomalies of tumor cells (ERBB2 copy number, presence of PIK3CA activating mutations) have been demonstrated to lie beyond resistance to HER2-targeting agents (22,23). Curtis’ team has recently shown, by using GeoMx® DSP on pre-, on-, and post-treatment HER2+ breast tumor tissue samples from the neoadjuvant TRIO US-B07 trial and a panel of 40 proteins involved in tumor biology and the immune response, that patients who reached the complete pathological response to HER2-targeted treatment had an increase in immune markers such as CD45 and CD8 and exhibited higher protein heterogeneity in tumor cell biomarkers than in the TME immune markers (24).
Treatment of HER2+ breast cancers has recently integrated HER2-ADCs into the therapeutic armamentarium, after impressive efficacy of these drugs observed in the DESTINY-Breast trials (25). Another ADC, sacituzumab govitecan, designed to bind TROP-2, has been recently placed in the first-line therapy of metastatic HER2- breast cancers (26). Although these drugs induced significant tumor reduction in a much greater number of patients than the drugs previously used in the same indication, a part of the treated tumors showed resistance. It is logical to hypothesize that ADC efficacy is influenced by ITH, both of the antibody-binding protein expression and intrinsic sensitivity to the cytotoxic payload of various areas or cell population within a tumor. HER2 protein expression ITH has been demonstrated to influence accuracy of HER2+ tumor diagnosis and the efficacy of HER2-targeting antibody-based drugs such as trastuzumab (27). Furthermore, heterogeneity of HER2-expressing breast cancers has been demonstrated at the genomic level as well, both in the ERBB2-amplified and the non-amplified subcategory (28), where the latter is nowadays called HER2-low if the IHC score for HER2 expression is 2+ or 1+ (29). In translational studies done on tissue samples collected in several recent trials of HER2-ADCs in breast cancer, decreased level of HER2 protein expression or a high number of cells with IHC HER2 score 0 were highlighted as potential biomarker of resistance to these drugs, however the number of samples analyzed was small so the observations should be validated in larger cohorts (30).
In addition to the HER2 protein level expression in the tumor cells, the proficient immune response and the corresponding TME composition have been shown to predict good response to anti-HER2 drugs in breast and other cancers (31). On the basis of findings in other cancers (32), we can hypothesize that proximity of the tumor cells and the TME cells activated in the immune response would be important for good response to HER2-ADCs. Similarly, the spatial relationship between the immune TME, intratumor blood vessels, and tumor cells seems to be a plausible potential spatial biomarker of response to ADCs. These hypotheses remain to be verified as well as the one treating individual tumor cell sensitivity to payloads delivered by ADCs and the global tumor sensitivity to these drugs. Technologies that can resolve molecular expression at the single-cell level would be of great help in answering these questions (33).
Spatial biomarkers
By revealing fine details of ITH, technologies for spatially resolved molecular expression analysis gave birth to a new group of biomarkers, the spatial biomarkers. These biomarkers are combinations of at least one molecular expression and one specific localization within a tissue or a cell. As such, they belong to the composite biomarkers. Some of them are still basically qualitative, whereas some are expressed as scores, such as the Mixing Score, Immune HotSpot Score, or Tumor Budding/T-Cell Score (34). Another type of spatial biomarkers would be a “niche”, a group of cells having a particular pheno- or genotype, interacting between each other and having a specific localization within tumor tissue (35).
The growing number of spatial biomarkers is calling for at least three things: (I) their adaptation for clinical practice and robust validation both of ‘pure’ spatial parameters and of combined spatial-and-traditional (clinico-pathological) parameters; (II) improvement of tissue sampling, on the basis of spatial tissue analyses: where and how to sample a 3D structure such as a malignant tumor, to cover all aspects of ITH; and (III) development of the “From-Many-To-Few” approach, to facilitate implementation of newly-discovered spatial biomarkers into translational studies and, in fine, in clinical practice (36).
Conclusions
We are witnessing an intense development of spatial omics methods and technologies, which represent, first and foremost, exquisitely valuable tools for explorative tissue and cell investigations.
In the coming years, a number of ADCs and RICs will likely get approved for a tissue-agnostic use, but this will not reduce the need for deep exploration of tumor tissues before, during, and after treatment by these drugs. For example, the differences in organ immunology, impacting the composition of TME, might lie behind differences in ADC activity in different tumors. Therefore, translational studies associated with clinical trials of ADC/RIC-based treatments should integrate spatial omics, and the generated spatial data should be available to the scientific and medical community, for comparisons between trials, organs, or patient populations. Combination of spatial tissue analyses with molecular imaging and intratumor pharmacological assessments will likely be crucial for improvement of ADC/RIC specificity and efficacy as well as for patient selection based on integrative prediction of their response.
Acknowledgments
Funding: None.
Footnote
Peer Review File: Available at https://tbcr.amegroups.com/article/view/10.21037/tbcr-23-38/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tbcr.amegroups.com/article/view/10.21037/tbcr-23-38/coif). FPL receives Honoraria payment from Astra Zeneca. NRR receives Honoraria payment from MSD and Novartis and was supported by NanoString Technology. The other author has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zinn S, Vazquez-Lombardi R, Zimmermann C, et al. Advances in antibody-based therapy in oncology. Nat Cancer 2023;4:165-80. [Crossref] [PubMed]
- Marusyk A, Janiszewska M, Polyak K. Intratumor Heterogeneity: The Rosetta Stone of Therapy Resistance. Cancer Cell 2020;37:471-84. [Crossref] [PubMed]
- Goltsev Y, Samusik N, Kennedy-Darling J, et al. Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell 2018;174:968-981.e15. [Crossref] [PubMed]
- Black S, Phillips D, Hickey JW, et al. CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat Protoc 2021;16:3802-35. [Crossref] [PubMed]
- Lin JR, Izar B, Wang S, et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. Elife 2018;7:e31657. [Crossref] [PubMed]
- Bady E, Möller K, Mandelkow T, et al. BLEACH&STAIN 15-marker Multiplexed Imaging in 3,098 Human Carcinomas Reveals Six Major PD-L1-driven Immune Phenotypes with Distinct Spatial Orchestration. Mol Cancer Res 2023;21:605-13. [Crossref] [PubMed]
- Hsieh WC, Budiarto BR, Wang YF, et al. Spatial multi-omics analyses of the tumor immune microenvironment. J Biomed Sci 2022;29:96. [Crossref] [PubMed]
- Park J, Kim J, Lewy T, et al. Spatial omics technologies at multimodal and single cell/subcellular level. Genome Biol 2022;23:256. [Crossref] [PubMed]
- Baysoy A, Bai Z, Satija R, et al. The technological landscape and applications of single-cell multi-omics. Nat Rev Mol Cell Biol 2023;24:695-713. [Crossref] [PubMed]
- Vandereyken K, Sifrim A, Thienpont B, et al. Methods and applications for single-cell and spatial multi-omics. Nat Rev Genet 2023;24:494-515. [Crossref] [PubMed]
- Huyghe N, Benidovskaya E, Beyaert S, et al. Multiplex Immunofluorescence Combined with Spatial Image Analysis for the Clinical and Biological Assessment of the Tumor Microenvironment. J Vis Exp 2023; [Crossref] [PubMed]
- Chang Q, Ornatsky OI, Siddiqui I, et al. Imaging Mass Cytometry. Cytometry A 2017;91:160-9. [Crossref] [PubMed]
- Keren L, Bosse M, Thompson S, et al. MIBI-TOF: A multiplexed imaging platform relates cellular phenotypes and tissue structure. Sci Adv 2019;5:eaax5851. [Crossref] [PubMed]
- Bassiouni R, Gibbs LD, Craig DW, et al. Applicability of spatial transcriptional profiling to cancer research. Mol Cell 2021;81:1631-9. [Crossref] [PubMed]
- Williams CG, Lee HJ, Asatsuma T, et al. An introduction to spatial transcriptomics for biomedical research. Genome Med 2022;14:68. [Crossref] [PubMed]
- Park HE, Jo SH, Lee RH, et al. Spatial Transcriptomics: Technical Aspects of Recent Developments and Their Applications in Neuroscience and Cancer Research. Adv Sci (Weinh) 2023;10:e2206939. [Crossref] [PubMed]
- He S, Bhatt R, Brown C, et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat Biotechnol 2022;40:1794-806. [Crossref] [PubMed]
- Merritt CR, Ong GT, Church SE, et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat Biotechnol 2020;38:586-99. [Crossref] [PubMed]
- Zhang D, Deng Y, Kukanja P, et al. Spatial epigenome-transcriptome co-profiling of mammalian tissues. Nature 2023;616:113-22. [Crossref] [PubMed]
- Toki MI, Merritt CR, Wong PF, et al. High-Plex Predictive Marker Discovery for Melanoma Immunotherapy-Treated Patients Using Digital Spatial Profiling. Clin Cancer Res 2019;25:5503-12. [Crossref] [PubMed]
- Hernandez S, Das P, Holliday EB, et al. Differential Spatial Gene and Protein Expression Associated with Recurrence Following Chemoradiation for Localized Anal Squamous Cell Cancer. Cancers (Basel) 2023;15:1701. [Crossref] [PubMed]
- Janiszewska M, Stein S, Metzger Filho O, et al. The impact of tumor epithelial and microenvironmental heterogeneity on treatment responses in HER2+ breast cancer. JCI Insight 2021;6:e147617. [Crossref] [PubMed]
- Filho OM, Viale G, Stein S, et al. Impact of HER2 Heterogeneity on Treatment Response of Early-Stage HER2-Positive Breast Cancer: Phase II Neoadjuvant Clinical Trial of T-DM1 Combined with Pertuzumab. Cancer Discov 2021;11:2474-87. [Crossref] [PubMed]
- McNamara KL, Caswell-Jin JL, Joshi R, et al. Spatial proteomic characterization of HER2-positive breast tumors through neoadjuvant therapy predicts response. Nat Cancer 2021;2:400-13. [Crossref] [PubMed]
- Makhlin I, DeMichele A. Trastuzumab deruxtecan: An antibody-drug conjugate embracing its destiny in breast cancer. Cell Rep Med 2022;3:100668. [Crossref] [PubMed]
- Shastry M, Jacob S, Rugo HS, et al. Antibody-drug conjugates targeting TROP-2: Clinical development in metastatic breast cancer. Breast 2022;66:169-77. [Crossref] [PubMed]
- Nitta H, Li Z. Breast HER2 Intratumoral Heterogeneity as a Biomarker for Improving HER2-Targeted Therapy. Crit Rev Oncog 2020;25:233-40. [Crossref] [PubMed]
- Schettini F, Prat A. Dissecting the biological heterogeneity of HER2-positive breast cancer. Breast 2021;59:339-50. [Crossref] [PubMed]
- Schettini F, Chic N, Brasó-Maristany F, et al. Clinical, pathological, and PAM50 gene expression features of HER2-low breast cancer. NPJ Breast Cancer 2021;7:1. Erratum in: NPJ Breast Cancer 2023;9:32. [Crossref] [PubMed]
- Giugliano F, Carnevale Schianca A, Corti C, et al. Unlocking the Resistance to Anti-HER2 Treatments in Breast Cancer: The Issue of HER2 Spatial Distribution. Cancers (Basel) 2023;15:1385. [Crossref] [PubMed]
- Luque M, Sanz-Álvarez M, Morales-Gallego M, et al. Tumor-Infiltrating Lymphocytes and Immune Response in HER2-Positive Breast Cancer. Cancers (Basel) 2022;14:6034. [Crossref] [PubMed]
- Antoranz A, Van Herck Y, Bolognesi MM, et al. Mapping the Immune Landscape in Metastatic Melanoma Reveals Localized Cell-Cell Interactions That Predict Immunotherapy Response. Cancer Res 2022;82:3275-90. [Crossref] [PubMed]
- Ahmed R, Zaman T, Chowdhury F, et al. Single-Cell RNA Sequencing with Spatial Transcriptomics of Cancer Tissues. Int J Mol Sci 2022;23:3042. [Crossref] [PubMed]
- Fomitcheva-Khartchenko A, Kashyap A, Geiger T, et al. Space in cancer biology: its role and implications. Trends Cancer 2022;8:1019-32. [Crossref] [PubMed]
- Theis FJ, Dar D, Vento-Tormo R, et al. What do you most hope spatial molecular profiling will help us understand? Part 1. Cell Syst 2023;14:423-7. [Crossref] [PubMed]
- Akhoundova D, Rubin MA. Clinical application of advanced multi-omics tumor profiling: Shaping precision oncology of the future. Cancer Cell 2022;40:920-38. [Crossref] [PubMed]
Cite this article as: Radosevic-Robin N, Kossai M, Penault-Llorca F. New-generation technologies for spatial tissue analysis, indispensable tools for deciphering intratumor heterogeneity in the development of antibody-drug conjugates and radio-immunoconjugates for cancer treatment. Transl Breast Cancer Res 2023;4:28.


