
Biomarkers derived from CmP signal network in triple negative breast cancers
Introduction
Breast cancer is frequently detected and ranks second in terms of causing death among women due to cancer (1,2). It constitutes around 30% of all newly diagnosed cancer cases (3-8). The survival rates of breast cancer are greatly influenced by the stage at which it is diagnosed, with early detection leading to higher chances of survival (3,5,7). Multiple factors such as family history, alcohol consumption, and hormone exposure play a role in the development of breast cancer (9-11). Triple-negative breast cancer (TNBC) is a highly aggressive and heterogeneous form of breast cancer, distinguished by the absence of three prominent receptors: estrogen receptor (ER), classic nuclear progesterone receptor (nPR), and receptor tyrosine-protein kinase erbB-2 (HER2) (12,13). TNBC is known for its significant heterogeneity, encompassing various histological, molecular, and immune subtypes that contribute to distinct clinical outcomes that are still not fully understood (14-16). Among TNBCs, over 70% are of basal phenotype, one of the most aggressive TNBCs, and non-classic membrane progesterone receptors (mPRs) role in tumorigenesis, especially in basal phenotype, is of great interest since basal phenotype-derived breast cancer cells do not express nPRs (17). The cerebral cavernous malformation signaling complex (CSC)-mPR-progesterone (PRG) (CmP)/CSC-mPR-PRG-nPR (CmPn) signaling network, which integrates the CSC with mPRs and nPRs, plays a role in breast cancer tumorigenesis. Understanding these pathways can provide insights into potential treatments. The involvement of mPRs in tumorigenesis is still under study, with initial beliefs suggesting their lack of genomic actions, but recent research has identified PRG receptors (PRs) throughout the cell. This paper aims to provide a comprehensive overview of the CmP/CmPn pathways and their significance in TNBC.
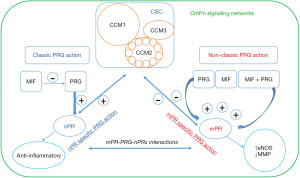
Cerebral cavernous malformations (CCMs) are abnormal dilations of small blood vessels in the brain, increasing the risk of stroke (18,19). Familial CCMs are hereditary and follow an autosomal dominant pattern (18-23). Deficiency of CCM1, CCM2, and CCM3 proteins, resulting from loss-of-function genetic mutations, contributes to this condition by disrupting the CSC (Figure 1) (20-23). Initially, PRG was thought to have a primary role in preparing the endometrium (24-26). Current understanding has reached a consensus that PRG can exert its effects through both classic and non-classic pathways, influencing the progression of breast cancer during tumorigenesis. It can independently bind to classic nuclear PRG receptors (nPRs) or non-classic mPRs, activating parallel signaling cascades (24-32). The CSC can modulate the interaction between mPRs and nPRs, merging these pathways into CmPn signal networks (Figure 2) (33). These signal networks are relevant in other types of tumorigenesis and vascular malformation during angiogenesis (8,33-40). The feedback regulation between PRG-mediated signaling and the CSC signaling within the CmPn signal network plays a crucial role in breast cancer development (41). Long-term exposure to PRG is associated with increased breast cancer risk (10), highlighting the significance of this network in tumorigenesis (8,33-40). Considering the influence of factors on the CmPn signaling network, various elements like trace minerals, antagonists, and agonists for PRs (mPRs/nPRs) have been considered for their potential application in breast cancer prevention and treatment (8,33-36,42-44).

This review aims to provide a concise overview of the signaling networks linked to the CmP/CmPn pathways and their involvement in the development of TNBC. Additionally, it presents an evolving understanding of the role of mPRs in tumorigenesis (8,33-36,42-45).
Potential roles of CCM proteins in tumorigenesis
CCMs are characterized by abnormal dilation of small blood vessels in the brain, increasing the risk of stroke (18,19). This condition can be hereditary, following an autosomal dominant pattern of inheritance, and is associated with specific genes such as CCM1, CCM2, and CCM3 (18-23). These genes encode proteins that interact with each other, forming the CSC (20-23). While the majority of research on CCMs has centered around their impact on cerebral cavernous hemangiomas and the increased risk of hemorrhagic stroke (20-23), there is emerging evidence indicating a potential involvement of CCM genes in tumor formation of major types of cancers (37). CCM1, one of the CCM genes, exhibits expression in various tissue types, indicating its diverse contribution to cellular physiology and its broader involvement in tumorigenesis across multiple tissues. This suggests that the CSC may play a role in tumorigenesis. While the full extent of these roles requires more research, there is strong suspicion that they significantly contribute to reproductive cancers, as evidenced by the increased expression of CCM1 and CCM3 correlating with higher severity and stages of endometrial tumors (37). The altered expression of CCM genes in liver disease and cancer, such as CCM1 in early liver cirrhosis/hepatitis and stage one liver tumors, indicates a complex role in liver tumor development (37). Other members of the CSC, like CCM2, have also been observed in different types of tissues and cancers (46), including lymphomas, highlighting the intricate role of the CSC in tumorigenesis that requires further elucidation (37). These findings are not unexpected, as studies have revealed that growing CCMs often coincide with mutations in the PIK3CA gene, which is commonly associated with cancer (47,48). The correlation between angiogenesis, shared genes implicated in tumorigenesis (47,48), and CCMs has provided insights into the possible role of CmPn signaling network in the development of both diseases.
Inception of CmPn signaling networks
Originally, PRG was thought to primarily play a role in female reproductive systems, specifically in the preparation of the endometrium throughout pregnancies (24-26). However, current consensus acknowledges that PRG can influence breast cancer progression (27,29-32). It can independently bind to nPRs or mPRs, activating parallel signaling cascades (27-32). Previously, the CSC, which regulates microvascular integrity, was regarded as a distinct entity (20-23). However, recent studies have uncovered a connection between these formerly independent signaling pathways, as the CSC has the ability to modulate the interaction between mPRs and nPRs (33). This finding has led to the integration of these three “independent” signaling cascades, giving rise to the complexes referred to as CmPn signal network (Figure 2) (33-36). This network is present in both nPR-positive cells, providing a comprehensive framework for understanding the interconnectedness of these pathways (8,33-40,49). The comprehension of mPRs’ role in tumorigenesis has undergone an evolving process, in contrast to the well-established classic nPRs. Initially, it was proposed that mPRs are a novel type of PRs that does not elicit genomic actions. However, several studies have identified PRs in different cellular locations, including the nucleus (41). The CmPn signaling network has been proven to play a crucial role in breast cancer development. Long-term exposure to PRG has been associated with an increased risk of breast cancer (10), further emphasizing the significance of this network in tumorigenesis (8,33-40). The discovery of the role of CmPn signaling network and tumorigenesis has sparked further investigation into their potential role in early detection. In summary, this section provides an overview of the present understanding of the signaling networks related to the CmPn network. It investigates the role of these signaling pathways in the development of TNBC and discusses potential applications for future treatment strategies.
Factors influencing CmPn/CmP signaling networks
Considering the influence of some key factors on the CmPn signaling network, various elements like trace minerals, antagonists, and agonists for PRs for their potential application in breast cancer prevention and treatment. This section aims to provide a brief summary of the factors associated with the CmPn network, the shift from CmPn to CmP network in nPR(−) TNBC cells, and the significance of the CmP network in TNBC development. Furthermore, we will examine the possible application of these factors in forthcoming treatment strategies.
Mifepristone has demonstrated potential in reducing the growth of TNBC cell lines (42-45), which can be partly attributed to its mechanism of action within the CmP signaling pathway (8). Acting as both an antagonist for nPRs and an agonist for mPRs (42-45), mifepristone has shown the ability to suppress basal TNBC stem cells and inhibit the growth of both nPR(+/−) and nPR(−) TNBC cells in the presence of elevated levels of PRG (8,33-36,42-44). It is important to note that in nPR(−) TNBC cells, the combined effects of mifepristone and PRG specifically target mPRs, working synergistically on these receptors. Using mifepristone to target breast cancer stem cells in TNBC has been identified as a treatment approach, opening up possibilities for exploring other functions within the CmP signaling cascade that could be targeted in future treatments (41,45). The ongoing investigation of the CmP pathway provides hope for the development of innovative therapeutic strategies for TNBC.
Zinc, an essential mineral with diverse physiological functions, plays a critical role in activating numerous enzymes in the body (8). It is involved in processes such as DNA synthesis, regulation of the female reproductive system, and maintenance of the immune system (50). Recent research has investigated the complex relationship between zinc and breast cancer development. Reduced zinc consumption has been linked to an increased risk of breast cancer, likely due to low levels of zinc between cells and elevated levels within cells (41). In the case of mutated mPRs, higher intracellular zinc levels facilitate the binding of PRG to mPRs, thereby restoring PRG’s ability to exert its effects within the cell (51). This finding provides insights into breast cancers that lack nPRs and highlights how PRG can still induce cellular mutations. The evidence of intracellular zinc enabling PRG binding through mPRs to influence CmP signaling network, presents challenges in the treatment of all nPR-negative cancers, particularly TNBC (8).
Role of CmP in TNBC
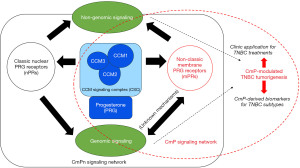
TNBC is recognized as one of the most lethal types of breast cancer due to its aggressive nature and the absence of ER, PR, and HER2 expression (12,13). TNBC displays substantial heterogeneity, involving diverse histological, molecular, and immune subtypes that contribute to distinct clinical outcomes that are not yet fully comprehended (14-17). TNBC poses challenges for treatment due to its aggressive nature and lack of ER, nPRs, and HER2 expression (34,35). However, recent findings have shown that TNBC still interacts with PRG through non-classic membrane receptors (34,35). This discovery led to the identification of the novel hormone route known as the CmP, where PRG utilizes mPRs and the CSC to deliver PRG to the nuclear membrane (Figure 3) (34,35,41). Further research has confirmed the higher abundance of mPRs in TNBC compared to cancers with nPRs (34,35). This suggests that TNBC may receive continuous PRG signaling, creating an environment favorable for rapid metastasis (8,33-36,38-40). These findings have also been supported by treatment options that have shown greater success in TNBC.
Biomarkers discovered through CmP in TNBC
Breast cancers exhibit significant clinical heterogeneity, and this variability is particularly prominent in TNBCs (52-55). TNBCs exhibit distinct variations in tumor aggressiveness, relapse rates, response to endocrine therapy, and sensitivity to cytotoxic chemotherapy (56-58). Hence, it is crucial to identify specific prognostic and predictive biomarkers unique to TNBC subtypes to guide clinical decision-making (59-62). Biomarkers, encompassing objectively measurable characteristics, play a vital role in predicting, diagnosing, prognosing, and assessing disease progression, regression, and treatment outcomes. Lehmann’s proposed sub-classification of TNBCs has demonstrated variability, ranging from seven to four subtypes, each with advantages and disadvantages based on the context Technological advancements and expression profiling have significantly contributed to a more comprehensive characterization of TNBC subgroups, leading to the identification of precise biomarkers, therapeutic targets, and a better understanding of the underlying molecular mechanisms associated with TNBC (63-67). Tumor immune interactions in TNBCs are intricate and heterogeneous, influenced by diverse gene expressions within the tumor immune microenvironment. Various immune subtypes have been identified, impacting tumor-immune interactions and patient survival (68). Numerous biomarkers and signatures tailored to TNBC subtypes, addressing the challenges of immunotherapy, have been reported with prognostic value (16,69-73). Early detection of breast cancer is crucial for saving lives, and diagnostic and prognostic biomarkers play a pivotal role in achieving this goal (71,74-77). Biomarkers provide valuable information regarding cancer staging, location, and signaling cascades involved in cancer development (38,78-82). The discovery of novel biomarkers continues to advance our understanding of TNBC and holds promise for improved diagnosis and treatment. While numerous biomarkers have been identified, it is important to recognize the potential of prognostic and etiological biomarkers for early detection and as targets for treatment options (34,83,84). These biomarkers hold the key to unlocking pathways that can be utilized for developing effective therapies (33-35,37,38).
Alternative options
The current treatment options for TNBC are limited to chemotherapy, targeted agents, and immune checkpoint inhibitors, with chemotherapy being the standard approach (85). However, even with the introduction of targeted agents and immune checkpoint inhibitors, the overall survival rate for metastatic TNBC beyond 5 years remains at only 11% (86). In order to improve this outcome, it is crucial to explore alternative signaling cascades such as the CmP pathway, which holds potential for novel treatment strategies that have yet to be investigated (33-35,37,38). Notably, changes in CCM gene expression have been observed in reproductive cancers, including breast cancer, indicating a potential avenue for preventing tumor metastasis, which is associated with poor prognosis (34,35,37). Further research and understanding of the mechanisms involved in the CmP signaling cascade could lead to the identification of additional targeted therapies for future treatments, offering hope for improved outcomes in TNBC.
Investigating biomarkers derived from the CmP signal network holds promise for TNBC diagnosis and treatment. By analyzing the components of this network, researchers are able to identify specific molecular markers that can distinguish TNBC from other breast cancer subtypes. These biomarkers can provide valuable information about the prognosis and potential therapeutic targets for TNBC patients (34,35).
Additionally, understanding the dynamics and interactions within the CmP signal network can help identify novel therapeutic approaches. By targeting specific components of this network, such as mPRs or CSC, researchers may develop personalized treatments for TNBC patients. This approach could potentially improve patient outcomes and reduce the aggressiveness of this subtype.
Conclusions
This review focuses on the development of diagnostic and prognostic biomarkers in breast cancer, particularly in the context of TNBC. It highlights the importance of understanding the CmP signaling cascade and its associated factors in breast cancer tumorigenesis. The discovery of mPRs and the involvement of the CSC have provided new insights into the mechanisms underlying breast cancer progression. These findings contribute to the identification of prognostic biomarkers and the development of targeted therapies that interact with the CmP signaling cascade and its key components. The review emphasizes the potential of targeting various components of the CmP network, such as mPRs or CCM proteins, and utilizing biomarker-guided approaches to improve the prognosis and life expectancy of breast cancer patients and those with related conditions.
In summary, the exploration of biomarkers derived from the CmP signal network in TNBCs holds great promise for advancing our understanding of this aggressive subtype of breast cancer. By deciphering the intricate mechanisms within this network, researchers can potentially develop targeted therapies and improve patient outcomes in TNBC.
Acknowledgments
The authors thank Revathi Gnanasekaran [Department of Molecular and Translational Medicine (MTM), Texas Tech University Health Science Center El Paso, El Paso, TX, USA] and Odalys Quintanar [Department of Molecular and Translational Medicine (MTM), Texas Tech University Health Science Center El Paso, El Paso, TX, USA] for their kind help during the preparation of this article.
Funding: None.
Footnote
Peer Review File: Available at https://tbcr.amegroups.com/article/view/10.21037/tbcr-23-30/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tbcr.amegroups.com/article/view/10.21037/tbcr-23-30/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Baquet CR, Mishra SI, Commiskey P, et al. Breast cancer epidemiology in blacks and whites: disparities in incidence, mortality, survival rates and histology. J Natl Med Assoc 2008;100:480-8. [Crossref] [PubMed]
- Bauer KR, Brown M, Cress RD, et al. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer 2007;109:1721-8. [Crossref] [PubMed]
- Siegel RL, Miller KD, Wagle NS, et al. Cancer statistics, 2023. CA Cancer J Clin 2023;73:17-48. [Crossref] [PubMed]
- Wilkinson L, Gathani T. Understanding breast cancer as a global health concern. Br J Radiol 2022;95:20211033. [Crossref] [PubMed]
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Narayan AK, Lee CI, Lehman CD. Screening for Breast Cancer. Med Clin North Am 2020;104:1007-21. [Crossref] [PubMed]
- Giaquinto AN, Sung H, Miller KD, et al. Breast Cancer Statistics, 2022. CA Cancer J Clin 2022;72:524-41. [Crossref] [PubMed]
- Renteria M, Belkin O, Jang D, et al. CmPn signaling networks in the tumorigenesis of breast cancer. Front Endocrinol (Lausanne) 2022;13:1013892. [Crossref] [PubMed]
- Sun YS, Zhao Z, Yang ZN, et al. Risk Factors and Preventions of Breast Cancer. Int J Biol Sci 2017;13:1387-97. [Crossref] [PubMed]
- Trabert B, Sherman ME, Kannan N, et al. Progesterone and Breast Cancer. Endocr Rev 2020;41:320-44. [Crossref] [PubMed]
- Moore MR, Spence JB, Kiningham KK, et al. Progestin inhibition of cell death in human breast cancer cell lines. J Steroid Biochem Mol Biol 2006;98:218-27. [Crossref] [PubMed]
- Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 2006;295:2492-502. [Crossref] [PubMed]
- Seal MD, Chia SK. What is the difference between triple-negative and basal breast cancers? Cancer J 2010;16:12-6. [Crossref] [PubMed]
- Lehmann BD, Colaprico A, Silva TC, et al. Multi-omics analysis identifies therapeutic vulnerabilities in triple-negative breast cancer subtypes. Nat Commun 2021;12:6276. [Crossref] [PubMed]
- Asleh K, Riaz N, Nielsen TO. Heterogeneity of triple negative breast cancer: Current advances in subtyping and treatment implications. J Exp Clin Cancer Res 2022;41:265. [Crossref] [PubMed]
- Kudelova E, Smolar M, Holubekova V, et al. Genetic Heterogeneity, Tumor Microenvironment and Immunotherapy in Triple-Negative Breast Cancer. Int J Mol Sci 2022;23:14937. [Crossref] [PubMed]
- Zuo L, Li W, You S. Progesterone reverses the mesenchymal phenotypes of basal phenotype breast cancer cells via a membrane progesterone receptor mediated pathway. Breast Cancer Res 2010;12:R34. [Crossref] [PubMed]
- Batra S, Lin D, Recinos PF, et al. Cavernous malformations: natural history, diagnosis and treatment. Nat Rev Neurol 2009;5:659-70. [Crossref] [PubMed]
- Draheim KM, Fisher OS, Boggon TJ, et al. Cerebral cavernous malformation proteins at a glance. J Cell Sci 2014;127:701-7. [Crossref] [PubMed]
- Padarti A, Zhang J. Recent advances in cerebral cavernous malformation research. Vessel Plus 2018;2:21. [Crossref] [PubMed]
- Zhang J. Molecular biology of cerebral cavernous malformation. In: Rigamonti D. editor. Cavernous Malformations of the Nervous System. Cambridge: Cambridge University Press; 2011:31-40.
- Zhang J. Learn from the past, review the present, and look towards the future. Vessel Plus 2022;6:20.
- Liu H, Rigamonti D, Badr A, et al. Ccm1 regulates microvascular morphogenesis during angiogenesis. J Vasc Res 2011;48:130-40. [Crossref] [PubMed]
- Stouffer RL. Progesterone as a mediator of gonadotrophin action in the corpus luteum: beyond steroidogenesis. Hum Reprod Update 2003;9:99-117. [Crossref] [PubMed]
- Cable JK, Grider MH. Physiology, Progesterone. In: StatPearls. Treasure Island, FL, USA: StatPearls Publishing; 2023.
- Holesh JE, Bass AN, Lord M. Physiology, Ovulation. In: StatPearls. Treasure Island, FL, USA: StatPearls Publishing; 2023.
- Sleiter N, Pang Y, Park C, et al. Progesterone receptor A (PRA) and PRB-independent effects of progesterone on gonadotropin-releasing hormone release. Endocrinology 2009;150:3833-44. [Crossref] [PubMed]
- Pang Y, Thomas P. Progesterone signals through membrane progesterone receptors (mPRs) in MDA-MB-468 and mPR-transfected MDA-MB-231 breast cancer cells which lack full-length and N-terminally truncated isoforms of the nuclear progesterone receptor. Steroids 2011;76:921-8. [Crossref] [PubMed]
- Karteris E, Zervou S, Pang Y, et al. Progesterone signaling in human myometrium through two novel membrane G protein-coupled receptors: potential role in functional progesterone withdrawal at term. Mol Endocrinol 2006;20:1519-34. [Crossref] [PubMed]
- Elsers DA, Masoud EM, Kamel NAMH, et al. Immunohistochemical signaling pathways of triple negative and triple positive breast cancers: What is new? Ann Diagn Pathol 2021;55:151831. [Crossref] [PubMed]
- Dressing GE, Thomas P. Identification of membrane progestin receptors in human breast cancer cell lines and biopsies and their potential involvement in breast cancer. Steroids 2007;72:111-6. [Crossref] [PubMed]
- Dressing GE, Alyea R, Pang Y, et al. Membrane progesterone receptors (mPRs) mediate progestin induced antimorbidity in breast cancer cells and are expressed in human breast tumors. Horm Cancer 2012;3:101-12. [Crossref] [PubMed]
- Abou-Fadel J, Jiang X, Grajeda B, et al. CCM signaling complex (CSC) couples both classic and non-classic Progesterone receptor signaling. Cell Commun Signal 2022;20:120. [Crossref] [PubMed]
- Abou-Fadel J, Bhalli M, Grajeda B, et al. CmP Signaling Network Leads to Identification of Prognostic Biomarkers for Triple-Negative Breast Cancer in Caucasian Women. Genet Test Mol Biomarkers 2022;26:198-219. [Crossref] [PubMed]
- Abou-Fadel J, Grajeda B, Jiang X, et al. CmP signaling network unveils novel biomarkers for triple negative breast cancer in African American women. Cancer Biomark 2022;34:607-36. [Crossref] [PubMed]
- Abou-Fadel J, Jiang X, Padarti A, et al. mPR-Specific Actions Influence Maintenance of the Blood-Brain Barrier (BBB). Int J Mol Sci 2022;23:9684. [Crossref] [PubMed]
- Abou-Fadel J, Qu Y, Gonzalez EM, et al. Emerging roles of CCM genes during tumorigenesis with potential application as novel biomarkers across major types of cancers. Oncol Rep 2020;43:1945-63. [Crossref] [PubMed]
- Abou-Fadel J, Reid V, Le A, et al. Key Members of the CmPn as Biomarkers Distinguish Histological and Immune Subtypes of Hepatic Cancers. Diagnostics (Basel) 2023;13:1012. [Crossref] [PubMed]
- Renteria M, Belkin O, Aickareth J, et al. Zinc's Association with the CmPn/CmP Signaling Network in Breast Cancer Tumorigenesis. Biomolecules 2022;12:1672. [Crossref] [PubMed]
- Aickareth J, Hawwar M, Sanchez N, et al. Membrane Progesterone Receptors (mPRs/PAQRs) Are Going beyond Its Initial Definitions. Membranes (Basel) 2023;13:260. [Crossref] [PubMed]
- Zhang J, Abou-Fadel JS. Calm the raging hormone - A new therapeutic strategy involving progesterone-signaling for hemorrhagic CCMs. Vessel Plus 2021;5:48.
- Liang Y, Hou M, Kallab AM, et al. Induction of antiproliferation and apoptosis in estrogen receptor negative MDA-231 human breast cancer cells by mifepristone and 4-hydroxytamoxifen combination therapy: a role for TGFbeta1. Int J Oncol 2003;23:369-80.
- Moe BG, Vereide AB, Orbo A, et al. High concentrations of progesterone and mifepristone mutually reinforce cell cycle retardation and induction of apoptosis. Anticancer Res 2009;29:1053-8.
- Fjelldal R, Moe BT, Ørbo A, et al. MCF-7 cell apoptosis and cell cycle arrest: non-genomic effects of progesterone and mifepristone (RU-486). Anticancer Res 2010;30:4835-40.
- Liu R, Shi P, Nie Z, et al. Mifepristone Suppresses Basal Triple-Negative Breast Cancer Stem Cells by Down-regulating KLF5 Expression. Theranostics 2016;6:533-44. [Crossref] [PubMed]
- Jiang X, Padarti A, Qu Y, et al. Alternatively spliced isoforms reveal a novel type of PTB domain in CCM2 protein. Sci Rep 2019;9:15808. [Crossref] [PubMed]
- Zhang J, Abou-Fadel J, Renteria M, et al. Cerebral cavernous malformations do not fall in the spectrum of PIK3CA-related overgrowth. J Neurol Neurosurg Psychiatry 2022;jnnp-2022-328901. [Crossref] [PubMed]
- Zhang J, Croft J, Le A. Familial CCM Genes Might Not Be Main Drivers for Pathogenesis of Sporadic CCMs-Genetic Similarity between Cancers and Vascular Malformations. J Pers Med 2023;13:673. [Crossref] [PubMed]
- Gnanasekaran R, Aickareth J, Hawwar M, et al. CmPn/CmP Signaling Networks in the Maintenance of the Blood Vessel Barrier. J Pers Med 2023;13:751. [Crossref] [PubMed]
- Bedwal RS, Bahuguna A. Zinc, copper and selenium in reproduction. Experientia 1994;50:626-40. [Crossref] [PubMed]
- Thomas P. Membrane Progesterone Receptors (mPRs, PAQRs): Review of Structural and Signaling Characteristics. Cells 2022;11:1785. [Crossref] [PubMed]
- Roulot A, Héquet D, Guinebretière JM, et al. Tumoral heterogeneity of breast cancer. Ann Biol Clin (Paris) 2016;74:653-60. [Crossref] [PubMed]
- Polyak K. Heterogeneity in breast cancer. J Clin Invest 2011;121:3786-8. [Crossref] [PubMed]
- Pal SK, Childs BH, Pegram M. Triple negative breast cancer: unmet medical needs. Breast Cancer Res Treat 2011;125:627-36. [Crossref] [PubMed]
- Metzger-Filho O, Tutt A, de Azambuja E, et al. Dissecting the heterogeneity of triple-negative breast cancer. J Clin Oncol 2012;30:1879-87. [Crossref] [PubMed]
- Podo F, Buydens LM, Degani H, et al. Triple-negative breast cancer: present challenges and new perspectives. Mol Oncol 2010;4:209-29. [Crossref] [PubMed]
- Damaskos C, Garmpi A, Nikolettos K, et al. Triple-Negative Breast Cancer: The Progress of Targeted Therapies and Future Tendencies. Anticancer Res 2019;39:5285-96. [Crossref] [PubMed]
- Abramson VG, Lehmann BD, Ballinger TJ, et al. Subtyping of triple-negative breast cancer: implications for therapy. Cancer 2015;121:8-16. [Crossref] [PubMed]
- Miquel-Cases A, Retèl VP, van Harten WH, et al. Decisions on Further Research for Predictive Biomarkers of High-Dose Alkylating Chemotherapy in Triple-Negative Breast Cancer: A Value of Information Analysis. Value Health 2016;19:419-30. [Crossref] [PubMed]
- Liu Y, Teng L, Fu S, et al. Highly heterogeneous-related genes of triple-negative breast cancer: potential diagnostic and prognostic biomarkers. BMC Cancer 2021;21:644. [Crossref] [PubMed]
- Lehmann BD, Pietenpol JA, Tan AR. Triple-negative breast cancer: molecular subtypes and new targets for therapy. Am Soc Clin Oncol Educ Book 2015;e31-9. [Crossref] [PubMed]
- Capobianco E. Overview of triple negative breast cancer prognostic signatures in the context of data science-driven clinico-genomics research. Ann Transl Med 2022;10:1300. [Crossref] [PubMed]
- Punetha A, Kotiya D. Advancements in Oncoproteomics Technologies: Treading toward Translation into Clinical Practice. Proteomes 2023;11:2. [Crossref] [PubMed]
- Li Y, Zhan Z, Yin X, et al. Targeted Therapeutic Strategies for Triple-Negative Breast Cancer. Front Oncol 2021;11:731535. [Crossref] [PubMed]
- Spratlin JL, Cohen RB, Eadens M, et al. Phase I pharmacologic and biologic study of ramucirumab (IMC-1121B), a fully human immunoglobulin G1 monoclonal antibody targeting the vascular endothelial growth factor receptor-2. J Clin Oncol 2010;28:780-7. [Crossref] [PubMed]
- Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 2001;98:10869-74. [Crossref] [PubMed]
- Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature 2000;406:747-52. [Crossref] [PubMed]
- Thorsson V, Gibbs DL, Brown SD, et al. The Immune Landscape of Cancer. Immunity 2018;48:812-30.e14. [Crossref] [PubMed]
- Zhou D, Li M, Yasin MH, et al. The prognostic value and immune microenvironment association of AR in HER2+ nonmetastatic breast cancer. NPJ Breast Cancer 2023;9:30. [Crossref] [PubMed]
- Huang R, Wang H, Hong J, et al. A senescence-associated signature refines the classification of different modification patterns and characterization of tumor immune microenvironment infiltration in triple-negative breast cancer. Front Pharmacol 2023;14:1191910. [Crossref] [PubMed]
- Yu J, Guo Z, Wang L. Progress and Challenges of Immunotherapy Predictive Biomarkers for Triple Negative Breast Cancer in the Era of Single-Cell Multi-Omics. Life (Basel) 2023;13:1189. [Crossref] [PubMed]
- Pu S, Zhou Y, Xie P, et al. Identification of necroptosis-related subtypes and prognosis model in triple negative breast cancer. Front Immunol 2022;13:964118. [Crossref] [PubMed]
- Stewart RL, Matynia AP, Factor RE, et al. Spatially-resolved quantification of proteins in triple negative breast cancers reveals differences in the immune microenvironment associated with prognosis. Sci Rep 2020;10:6598. [Crossref] [PubMed]
- Fleisher B, Clarke C, Ait-Oudhia S. Current advances in biomarkers for targeted therapy in triple-negative breast cancer. Breast Cancer (Dove Med Press) 2016;8:183-97. [Crossref] [PubMed]
- Lu B, Natarajan E, Balaji Raghavendran HR, et al. Molecular Classification, Treatment, and Genetic Biomarkers in Triple-Negative Breast Cancer: A Review. Technol Cancer Res Treat 2023;22:15330338221145246. [Crossref] [PubMed]
- Smith L, Qutob O, Watson MB, et al. Proteomic identification of putative biomarkers of radiotherapy resistance: a possible role for the 26S proteasome? Neoplasia 2009;11:1194-207. [Crossref] [PubMed]
- Zhong G, Lou W, Shen Q, et al. Identification of key genes as potential biomarkers for triple-negative breast cancer using integrating genomics analysis. Mol Med Rep 2020;21:557-66. [Crossref] [PubMed]
- Bao C, Lu Y, Chen J, et al. Exploring specific prognostic biomarkers in triple-negative breast cancer. Cell Death Dis 2019;10:807. [Crossref] [PubMed]
- Cocco S, Piezzo M, Calabrese A, et al. Biomarkers in Triple-Negative Breast Cancer: State-of-the-Art and Future Perspectives. Int J Mol Sci 2020;21:4579. [Crossref] [PubMed]
- Isaacs J, Anders C, McArthur H, et al. Biomarkers of Immune Checkpoint Blockade Response in Triple-Negative Breast Cancer. Curr Treat Options Oncol 2021;22:38. [Crossref] [PubMed]
- Srivastava M, Eidelman O, Craig J, et al. Serum Biomarkers for Racial Disparities in Breast Cancer Progression. Mil Med 2019;184:652-7. [Crossref] [PubMed]
- Yadav BS, Chanana P, Jhamb S. Biomarkers in triple negative breast cancer: A review. World J Clin Oncol 2015;6:252-63. [Crossref] [PubMed]
- Lin SY, Xia W, Wang JC, et al. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci U S A 2000;97:4262-6. [Crossref] [PubMed]
- Li S, Li S, Sun Y, et al. The expression of β-catenin in different subtypes of breast cancer and its clinical significance. Tumour Biol 2014;35:7693-8. [Crossref] [PubMed]
- Bianchini G, De Angelis C, Licata L, et al. Treatment landscape of triple-negative breast cancer - expanded options, evolving needs. Nat Rev Clin Oncol 2022;19:91-113. [Crossref] [PubMed]
- Catalano A, Iacopetta D, Ceramella J, et al. New Achievements for the Treatment of Triple-Negative Breast Cancer. Appl Sci 2022;12:5554.
Cite this article as: Sanchez N, Harvey C, Vincent D, Croft J, Zhang J. Biomarkers derived from CmP signal network in triple negative breast cancers. Transl Breast Cancer Res 2023;4:21.


