
Analysis of tumor heterogeneity at the molecular level: a case report of efficacy separation in metastatic breast cancer
Introduction
Breast cancer is a major public health concern and affects tens of thousands of women worldwide each year. Characterization of the genetic landscape of pulmonary metastases in breast cancer is of high importance to identify potential targets for personalized therapy regimens. In this paper we found that there were differences between the two lesions using next-generation sequencing, which provided ideas and basis for future precise treatment. Such intratumor heterogeneity poses significant clinical challenges, including identification of emergence of resistance to therapy (1). Next-generation sequencing approaches have allowed higher resolution of intratumor heterogeneity and resulted in a better conceptual understanding of cancer evolution. Accumulating evidences are suggestive of a branched evolution model, such that early acquired mutations are present ubiquitously, whereas subsequent mutational events are found in confined regions (2-4). This case is written in accordance with the CARE reporting guideline (available at http://dx.doi.org/10.21037/tbcr-20-20).
Case presentation
Clinical history
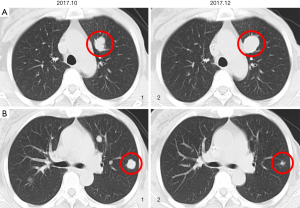
The patient, 51-year-old, underwent modified radical mastectomy for left breast cancer in December 2013. Postoperative pathology suggested grade III invasive ductal carcinoma, ER (++, 40%), PR (++, 60%), HER2 (−) and Ki67 (20%). Every 3 weeks for 3 cycles of AT [pirarubicin (60 mg 38 mg/m2 d1) combined with docetaxel (120 mg 75 mg/m2 d1)], every 3 weeks for 3 cycles of TX (docetaxel (120 mg 75 mg/m2 d1) combined with capecitabine (3,000 mg, 1,887 mg/m2 d1–14) and adjuvant endocrine therapy [tamoxifen (20 mg qd from August 2014 to December 2016), letrozole (2.5 mg qd from December 2016 to February 2017) and anastrozole (1 mg qd from February 2017 to April 2017), successively) were performed after surgery. Metastases to the bilateral lung, left chest wall and sternum were detected in April 2017. Subsequently, capecitabine (3,500 mg 2,108 mg/m2 d1–14) salvage therapy every 3 weeks from April 2017 to September 2017, toremifene (60 mg qd from September 2017 to October 2017) and albumin-bound paclitaxel (200 mg 120 mg/m2 d1, d8 every 3 weeks for 2 cycle from October 2017 to December 2017) salvage therapy was conducted. After 2 treatment cycles, the patient presented separation of efficacy in bilateral lung lesions, some lesions increased and some narrowed. Imaging showed that the largest metastatic lesion increased and the remaining lesions were significantly reduced (Figure 1). She underwent a left lung lesion resection and obtained two lung metastases lesions (one for drug-resistant lesions, one for sensitive lesions). Peripheral blood was obtained at the time of acquisition for lung tissue. The next-generation sequencing conducted on these samples. There was none of adverse and unanticipated outcomes. Besides, her mother had no breast cancer. To February 2020, the patient received salvage therapy with anastrozole (1 mg qd). The timeline of the patient in past 6 years in Figure 2.

Next-generation sequencing
Sample extraction
We performed whole genome sequencing of DNA from two pulmonary metastases, and normal tissue (whole blood) from a patient who had efficacy separation in metastatic breast cancer. DNA of tumor samples and DNA of blood sample were extracted using the KAPA HTP Library Preparation Kit and Life MagMAX Cell-Free DNA Isolation Kit respectively.
Library preparation and sequencing
Using the library preparation of KAPA Hyper Prep Kit to purify the DNA library, we need to accomplish the end-repair, addition of “A”, addition of linker and PCR reaction. The purified DNA library was enriched in the region of interest using the Anfutan gene detection. The enriched DNA samples were sequenced using the NextSeq500 next-generation sequencing platform, and the read length was 150 bp for double-end sequencing.
Sequencing data analysis
The BWA (Burrows Wheeler Aliger) software was used to compare with the human reference genome hg19. The repeats were labeled with Picard software. The contents of the BAM file were rearranged and corrected using the Genome Analysis Toolkit (GATK; Broad Institute), and the single nucleotide variation, insertion variation and deletion variation were identified using the Mutect2 tool under GATK software.
Traditional pathology
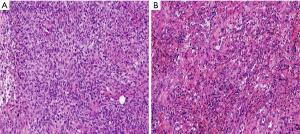
Histopathological analysis of these lesions revealed poorly differentiated sarcomatoid carcinoma (Figure 3). Immunohistochemical analysis of the two lesions from pulmonary metastases demonstrated that these lesions, in a way akin to the pulmonary metastases in breast cancer, were lacked ER, PR and HER2 expression. Based on these results, the histological and immunohistochemical finds suggested that these two lesions were not inconsistent.
Detection of immunohistochemical indicators
Biopsy specimens of primary lesions and recurrent metastases of breast cancer were fixed with 10% formaldehyde, embedded in paraffin, and sliced into 3-µm sections. The expressions of ER, PR, HER-2 and Ki-67 were detected by routine immunohistochemical S-P method. After dewaxing in xylene, the sections were hydrated by gradient alcohol, followed by antigen retrieval at high temperature and pressure. Corresponding primary antibody was then dripped, and the sections were washed three times using sulfate buffer (0.01 M, pH 7.5±0.1) for 3 min each time. Subsequently, the sections were dripped with secondary antibody DAC0 and developed with DAB. Afterwards, the sections were rinsed with distilled water, re-stained with alum hematoxylin for 5 min, differentiated with 0.2% hydrochloric acid, dehydrated with gradient alcohol, cleared in xylene and sealed with neutral gum. The observation was performed under an optical microscope.
Result reading
Each section was read by two experienced pathologists employing a double-blind method to draw conclusions. The known positive sections were used as positive controls and sulfate buffer (PBS) was used as negative control instead of primary antibody. Under a 400-fold microscope, 10 visual fields were randomly selected and the percentage of positive cells per 1,000 tumor cells was calculated. The results were evaluated according to the following criteria.
ER and PR
Both ER and PR were localized in the nucleus, and their positive expressions were determined by the presence of brown-yellow granules in the nucleus. According to positive cell count, the expression rate was divided into four grades: (−), the percentage of positive cells <10%; (+), the percentage of positive cells =10–25%; (++), the percentage of positive cells =25–50%; and (+++), the percentage of positive cells >50%.
HER-2
The positive-staining signals of HER-2 protein were localized in the cell membrane or cytoplasm, presenting fishnet shape under a high-power microscope. The presence of brown-yellow granules in the cell membrane or both cell membrane and cytoplasm was considered as positive staining, but cytoplasmic staining alone was not considered as positive staining. The staining intensity was divided into three grades: grade 1, light brown, grade 3, dark brown, grade 2, between grade 1 and grade 3. Based on the staining intensity, the expression rate was divided into four grades: (−), completely negative; (+), invasive tumor cells at any proportion showed weak and incomplete membranous staining or less than 10% tumor cells showed weak and complete membranous staining with staining intensity of grade 1; (++), more than 10% tumor cells showed weak or inconsistently complete membranous staining, or 30% tumor cells showed strong and complete membranous staining, with staining intensity of grade 2; (+++), the ratio of invasive tumor cells showing strong and complete membranous staining more than 30%, and staining intensity was grade 3.
Ki67
Ki67 staining was determined based on the percentage of fine brown-yellow granules in the nucleus. Ki67-positive granules were distributed in the nucleus. Brown granules in the nucleus were considered as positive staining.
Molecular pathology
The next-generation sequencing yielded average read depths ranging from 376× to 6,209× in the drug-resistant metastatic tumor sample, 572× to 3,937× in the drug-sensitive sample, and 340× to 2,213× in the circulating tumor sample. Depths of coverage over all mapped bases was >99.30% when considering the exome. The original sequencing depths of these three samples were at least 10,606×.
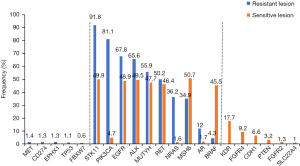
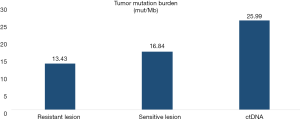
Gene mutations were detected in all three samples, involving 25 genes, including MUTYH, ALK, MSH6, PIK3CA, TP53, AR, BRAF, EGFR, EPHX1, NRAS, RET, STK11, FBXW7, CD274, MET. The genes in resistant lesion were: MUTYH, ALK, MSH6, PIK3CA, TP53, AR, BRAF, EGFR, EPHX1, NRAS, RET, STK11, FBXW7, CD274, MET. The genes in sensitive lesion were: FGFR4, MUTYH, ALK, MSH6, PIK3CA, TP53, AR, BRAF, EGFR, NRAS, RET, STK11, CDH1, FGFR2, SLC22A4, KDR, PTEN. The genes in blood sample were: FGFR4, MUTYH, ALK, MSH6, PIK3CA, TP53, AR, EGFR, EPHX1, NRAS, RET, STK11, FBXW7, FGFR2, SLC22A4, ABL1, CALR, DDR2, MYD88. Representation of the mutated genes in each specimen and their overlap (Table 1 and Figure 4). The tumor mutational burden of drug-resistant, sensitive and circulating tumor sample was 13.43, 16.84 and 25.99, respectively (Figure 5).

Full table
Discussion and conclusions
Characterization of the genetic landscape of pulmonary metastases in breast cancer is of high importance to identify potential targets for personalized therapy regimens. In the present study, two metastases samples of breast cancer were investigated with the next-generation sequencing to examine presence and extent of sub clonality within these rare metastatic lesions. Thereby, a profound genetic heterogeneity between two pulmonary metastases was revealed. This is the first case report, to our knowledge, of difference between lesions in lung metastases, suggesting the existence of a patient with distant metastases who would be different for therapy as a result of carrying heterogeneity.
There was no significant difference in the pathological results between the drug-resistant lesion and the sensitive lesion. Lung lesions were metastases of metaplastic breast carcinoma, which were diagnosed based on cell morphology and immunohistochemical results. Both lesions showed CK (+), suggesting that they originated from epithelial tissue. In addition, both lesions presented GATA-3 (+), suggesting their origin from breast tissue. Combined with the above results and the patient’s medical history, breast cancer metastases were diagnosed, rather than secondary primary tumors. Fibrosarcomatous components in metastatic lesions might be caused by the presence of metaplastic components in the primary lesions of breast cancer. However, due to surgical excision and pathological section preparation, these tissue components were not found. On the other hand, the cause might be that the primary lesions did not contain metaplastic components, but during the occurrence and progression of tumors, epithelial tissue re-differentiated into mesenchymal tissue, namely metaplastic components, after dedifferentiation.
Although both lesions were lung metastatic lesions of breast cancer, the expression of hormone receptor and HER2 receptor in primary and metastatic lesions were inconsistent. The causes for the inconsistency of hormone receptors between primary lesions and intrapulmonary metastases are as follows. Tumor heterogeneity Breast cancer is a highly heterogeneous malignant tumor (5). Even if they both are primary lesions, their difference in ER and PR expression may be caused by different sampling parts. With disease progression, the recurrent lesions may express different clinical and biological characteristics from primary lesions. Effects of systemic chemotherapy and endocrine therapy on tumor cells During systemic chemotherapy and endocrine therapy, tumor cells may change their signaling pathways, resulting in the difference in biological indicators between recurrent metastases and primary tumors (6). Changes in biological characteristics during tumor progression Because of the genetic instability of breast cancer (7), various biological characteristics may change during the occurrence and development tumors, which lead to different biological characteristics in recurrent metastases from primary lesions.
It was found that the drug-resistant lesions and the sensitive lesions have shared gene mutations and private gene mutations using the next-generation sequencing technology. The unique gene mutations of drug-resistant lesion were: CD274, MET, EPHX1, FBXW7 and TP53. Two genes of particular interest are CD274 and TP53, which were somatically mutated in the COSMIC database (8). Gene CD274 expresses CD274 protein (also known as PD-L1), which interacts with the PD-1 receptor on the surface of T cells, thereby inhibiting T cell activation, proliferation, and killing of tumor cells (9). Studies had found that the higher the tumor grade in the tumor tissues of patients with triple-negative breast cancer, the more PD-L1 expression, but no correlation with the prognosis (10). Other reports had found that high expression of PD-L1 in tumor cells was associated with poor prognosis of several malignancies such as non-small cell lung cancer (11), melanoma (12) and renal cancer (13). Muenst et al. (14) found that PD-L1 was expressed in 152 of 650 breast cancer tumor tissues, and breast cancer patients with PD-L1 expression had a worse prognosis, suggesting that the application of targeted therapy for PD-1/PD-L1 signaling pathway was of great significance. TP53 is a tumor suppressor which functions as a transcription factor and also plays a key role in the cellular response to stress (15). Germline mutations in TP53 causes Li-Fraumeni syndrome (16) and somatic mutations (17) are found in many human cancers. In this patient, the largest lesion increased after treatment with albumin paclitaxel, suggesting that the lesion was resistant to albumin paclitaxel. The unique gene mutations detected in this lesion may be related to the resistance of paclitaxel.
The mutation genes specific to sensitive lesions were: KDR, FGFR4, CDH1, FGFR2, PTEN and SLC22A4. Of particular interest is PTEN, which acts as a tumor suppressor by negatively regulating the AKT-PKB signaling pathway, and inhibits tumor development by antagonizing the activity of tyrosine kinase (18). Some researchers analyzed the PTEN heterozygosity, mutation profile and protein expression in 43 breast cancer patients and ten healthy individuals. The microsatellite markers used were D10S215, D10S541 and D10S579. As a result, it was found that there were mutations in each of exons 1, 5, 7 and 9 of the PTEN gene, and most of the mutations caused the PTEN protein to be truncated instead of lost activity. An increase in PTEN protein was detected in tissue specimens, suggesting that breast cancer may be associated with loss of PTEN protein activity rather than loss of expression (19). In addition, Rescigno et al. performed genetic testing on tumor tissues of 215 patients with prostate cancer who had failed castration therapy. Patients with PTEN mutations are more sensitive to docetaxel treatment (20). We found that the lesion was reduced by treatment with albumin paclitaxel, suggesting that the gene mutations KDR, FGFR4, CDH1, FGFR2, PTEN and SLC22A4 may be associated with paclitaxel.
In addition, Tumor mutational burden (TMB) can be used to quantitatively estimate the total number of base mutations in the tumor genome coding region. Tumor cells with higher levels of TMB are more likely to be recognized by the immune system, and may have a stronger immune response to a checkpoint inhibitor. Some tumor patients have mismatch repair defects, which are more likely to cause somatic mutations, resulting in increased tumor mutational load. Le et al. (21) found that patients with higher tumor mutational burden in colorectal cancer responded better to PD-1 inhibitor therapy, and then they further expanded the study to assessing the efficacy of PD-1 inhibitors in 12 different types of cancer patients (22), and found that most of the cancers with mismatch repair defects were more sensitive to PD-1 inhibitors. The tumor mutational burden of resistant lesion and sensitive lesion in this study were 13.4 and 16.8 respectively. The difference in tumor mutational load between drug-resistant lesion and sensitive lesion was not obvious.
Our data demonstrate a highly varying heterogeneity of pulmonary metastases in breast cancer. Due to the small number of cases, we can’t find the specific mutation gene that leads to the difference of treatment in the same patient. We hope that more cases will be sequenced in the future, and then find the key mutation gene. Moreover, tumor drivers of pulmonary metastases in breast cancer difference might not be detectable only by next-generation sequencing and further comprehensive studies including microsatellite instability as well as epigenetic analyses are needed.
Acknowledgments
Our sincere gratitude to the patient and her family for their permission to use the pictures for this paper. We thank Dr. Yanhong Tai for her pathological help. We gratefully acknowledge Dr. Juan Zhou for assistance with images. We also appreciate Wenjuan Huo for her interpretation of genetic testing report.
Funding: The next-generation sequencing was supported by the National Natural Science Foundation of China (No. 81472477) and the National Natural Science Foundation of China (No. 81602314). The funders provided financial support for genetic testing of the patient.
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist (available at http://dx.doi.org/10.21037/tbcr-20-20).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tbcr-20-20). ZJ serves as an unpaid Editor-in-Chief of Translational Breast Cancer Research. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). The patient has given the consent for the case report to be published.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 2015;27:15-26. [Crossref] [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. [Crossref] [PubMed]
- de Bruin EC, McGranahan N, Mitter R, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014;346:251-6. [Crossref] [PubMed]
- Yates LR, Gerstung M, Knappskog S, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 2015;21:751-9. [Crossref] [PubMed]
- Idirisinghe PK, Thike AA, Cheok PY, et al. Hormone receptor and c-ERBB2 status in distant metastatic and locally recurrent breast cancer. Pathologic correlations and clinical significance. Am J Clin Pathol 2010;133:416-29. [Crossref] [PubMed]
- Arslan C, Sari E, Aksoy S, et al. Variation in hormone receptor and HER-2 status between primary and metastatic breast cancer: review of the literature. Expert Opin Ther Targets 2011;15:21-30. [Crossref] [PubMed]
- Pusztai L, Viale G, Kelly CM, et al. Estrogen and HER-2 receptor discordance between primary breast cancer and metastasis. Oncologist 2010;15:1164-8. [Crossref] [PubMed]
- Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 2011;39:D945-50. [Crossref] [PubMed]
- Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:123-35. [Crossref] [PubMed]
- Guo L, Li W, Zhu X, et al. PD-L1 expression and CD274 gene alteration in triple-negative breast cancer: implication for prognostic biomarker. Springerplus 2016;5:805. [Crossref] [PubMed]
- Azuma K, Ota K, Kawahara A, et al. Association of PD-L1 overexpression with activating EGFR mutations in surgically resected nonsmall-cell lung cancer. Ann Oncol 2014;25:1935-40. [Crossref] [PubMed]
- Ott PA, Hodi FS, Robert C. CTLA-4 and PD-1/PD-L1 blockade: new immunotherapeutic modalities with durable clinical benefit in melanoma patients. Clin Cancer Res 2013;19:5300-9. [Crossref] [PubMed]
- Thompson RH, Dong H, Kwon ED. Implications of B7-H1 expression in clear cell carcinoma of the kidney for prognostication and therapy. Clin Cancer Res 2007;13:709s-715s. [Crossref] [PubMed]
- Muenst S, Schaerli AR, Gao F, et al. Expression of programmed death ligand 1 (PD-L1) is associated with poor prognosis in human breast cancer. Breast Cancer Res Treat 2014;146:15-24. [Crossref] [PubMed]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature 2000;408:307-10. [Crossref] [PubMed]
- Kamihara J, Rana HQ, Garber JE. Germline TP53 mutations and the changing landscape of Li-Fraumeni syndrome. Hum Mutat 2014;35:654-662. [Crossref] [PubMed]
- Petitjean A, Achatz MI, Borresen-Dale AL, et al. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene 2007;26:2157-65. [Crossref] [PubMed]
- Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol 2009;4:127-50. [Crossref] [PubMed]
- Kechagioglou P, Papi RM, Provatopoulou X, et al. Tumor suppressor PTEN in breast cancer: heterozygosity, mutations and protein expression. Anticancer Res 2014;34:1387-400. [PubMed]
- Rescigno P, Lorente D, Dolling D, et al. Docetaxel Treatment in PTEN- and ERG-aberrant Metastatic Prostate Cancers. Eur Urol Oncol 2018;1:71-7. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409-13. [Crossref] [PubMed]
Cite this article as: Li X, Bian L, Xu F, Qu Z, Yang F, Jiang Z. Analysis of tumor heterogeneity at the molecular level: a case report of efficacy separation in metastatic breast cancer. Transl Breast Cancer Res 2020;1:20.


